-
Notifications
You must be signed in to change notification settings - Fork 99
PredatorPrey_step5
This fifth step illustrates how to use parent species. Indeed, prey and predators share a few common features thus we will define a generic species that will regroup all the common elements (variables, behaviors, and aspects) between the prey and the predator species.
- Definition of a new generic species:
generic_species. - Definition of a new species:
predator. -
predatoragents move randomly. - At each simulation step, a
predatoragent can eat apreythat is localized at its grid cell.
We add four new parameters related to predator agents:
- The init number of predator agents.
- The max energy of the predator agents.
- The energy gained by a predator agent while eating a prey agent.
- The energy consumed by a predator agent at each time step.
We define four new global variables in the global section:
global {
...
int nb_predators_init <- 20;
float predator_max_energy <- 1.0;
float predator_energy_transfer <- 0.5;
float predator_energy_consum <- 0.02;
}
We define then the four corresponding parameters in the experiment:
parameter "Initial number of predators: " var: nb_predators_init min: 0 max: 200 category: "Predator" ;
parameter "Predator max energy: " var: predator_max_energy category: "Predator" ;
parameter "Predator energy transfer: " var: predator_energy_transfer category: "Predator" ;
parameter "Predator energy consumption: " var: predator_energy_consum category: "Predator" ;
A species can have a parent species: it automatically gets all the variables, skill and actions/reflex of the parent species.
We define a species called generic_species that is the parent of the species prey and predator:
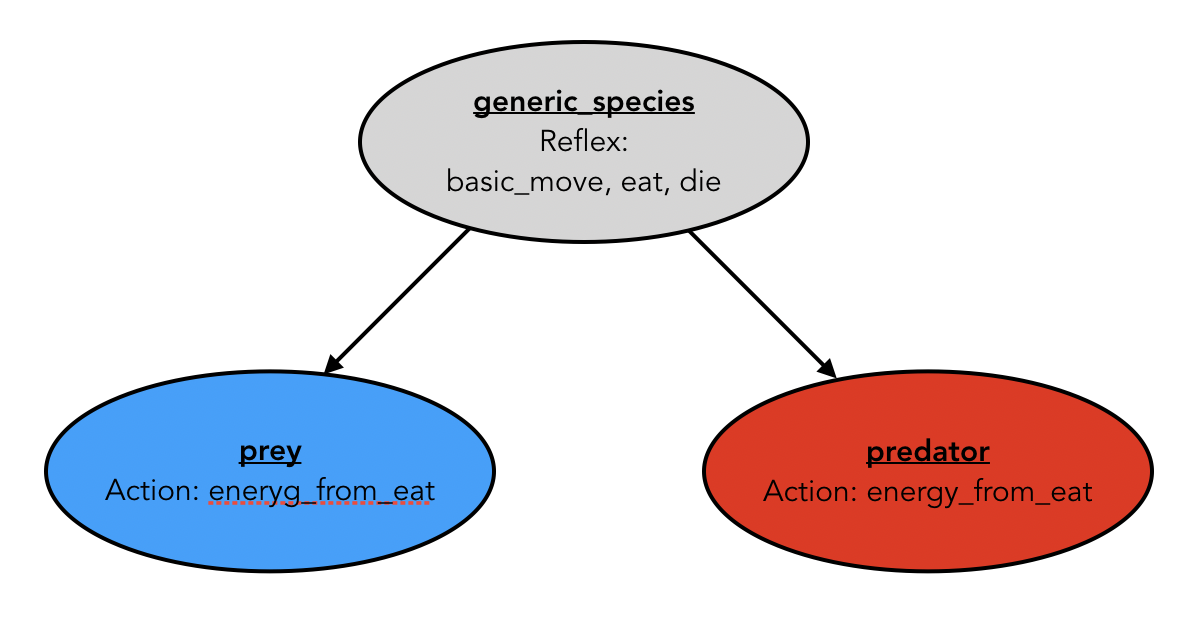
This species integrates all of the common feature between the prey and predator species:
- the variables:
sizecolormax_energymax_transferenergy_consummy_cellenergy
- the behaviors:
-
basic_movereflex -
eatreflex -
diereflex
-
- the actions:
-
energy_from_eataction
-
- the aspect:
-
baseaspect
-
As the eat behavior depends on the species (prey eats on vegetation_cell, whereas predator agents eat prey agents), we introduce an action energy_from_eat that will be redefined in each child species. Each species will implement its own eating behavior, returning the amount of energy it gets from this action.
We remind that an action is a capability available to the agents (what they can do). It is a block of statements that can be used and reused whenever needed.
- An action can accept arguments.
- An action can return a result (statement return).
There are two ways of calling an action: using a statement do or as part of an expression:
- for actions that do not return a result:
do action_name (arg1: v1 arg2: v2);
do action_name (v1, v2);
- for actions that return a result (which is stored in
my_var):
my_var <- action_name (arg1:v1, arg2:v2);
my_var <- action_name (v1, v2);
Thus the generic_species will have the following code. Note that the action energy_from_eat is also defined in this species, but with a default result (return 0.0).
species generic_species {
float size <- 1.0;
rgb color ;
float max_energy;
float max_transfer;
float energy_consum;
vegetation_cell my_cell <- one_of (vegetation_cell) ;
float energy <- rnd(max_energy) update: energy - energy_consum max: max_energy ;
init {
location <- my_cell.location;
}
reflex basic_move {
my_cell <- one_of (my_cell.neighbors2) ;
location <- my_cell.location ;
}
reflex eat {
energy <- energy + energy_from_eat();
}
reflex die when: energy <= 0 {
do die;
}
float energy_from_eat {
return 0.0;
}
aspect base {
draw circle(size) color: color ;
}
}
We specialize the prey species from the generic_species species:
- definition of the initial value of the agent variables.
- definition of the specific
eataction: if the current cell contains some food, the prey agent will take either all this food or themax_transfervalue (if the amount of food is greater than the maximum value the prey can take).
species prey parent: generic_species {
rgb color <- #blue;
float max_energy <- prey_max_energy ;
float max_transfer <- prey_max_transfer ;
float energy_consum <- prey_energy_consum ;
float energy_from_eat {
float energy_transfer <- 0.0;
if(my_cell.food > 0) {
energy_transfer <- min([max_transfer, my_cell.food]);
my_cell.food <- my_cell.food - energy_transfer;
}
return energy_transfer;
}
}
As done for the prey species, we specialize the predator species from the generic_species species:
- definition of the initial value of the agent variables.
- definition of the specific
eataction: first, the agent computes the list of prey agents contained bymy_cell(reachable_preysvariable); if it is not empty, it chooses one of the elements of this list, it kills it (i.e. asks it to die) and returns theenergy_transfervariable (that will be added to its own energy).
species predator parent: generic_species {
rgb color <- #red ;
float max_energy <- predator_max_energy ;
float energy_transfer <- predator_energy_transfer ;
float energy_consum <- predator_energy_consum ;
float energy_from_eat {
list<prey> reachable_preys <- prey inside (my_cell);
if(! empty(reachable_preys)) {
ask one_of (reachable_preys) {
do die;
}
return energy_transfer;
}
return 0.0;
}
}
Note that we used the ask statement. This statement allows to make a remote agent executes a list of statements.
Use of the ask statement as follows:
ask one_agent { }
or
ask agents_list { }
We used as well the species/agent list inside geometry/agent operator. This operator returns all the agents of the specified species (or from the specified agent list) that are inside the given geometry or agent geometry.
Like in the previous model, in the init block of the model, we create nb_predators_init.
global {
...
init {
create prey number: nb_preys_init ;
create predator number: nb_predators_init ;
}
}
Like in the previous model, we define a monitor to display the number of predator agents.
Definition of a global variable nb_predator that returns the current number of predator agents:
global {
...
int nb_predators -> {length (predator)};
...
}
Definition of the corresponding monitor:
monitor "number of predators" value: nb_predators ;
Also, do not forget to add the line to display predators in your simulation
display main_display {
...
species predator aspect: base ;
}
https://github.com/gama-platform/gama/blob/GAMA_1.9.2/msi.gama.models/models/Tutorials/Predator%20Prey/models/Model%2005.gaml
- Installation and Launching
- Workspace, Projects and Models
- Editing Models
- Running Experiments
- Running Headless
- Preferences
- Troubleshooting
- Introduction
- Manipulate basic Species
- Global Species
- Defining Advanced Species
- Defining GUI Experiment
- Exploring Models
- Optimizing Model Section
- Multi-Paradigm Modeling
- Manipulate OSM Data
- Diffusion
- Using Database
- Using FIPA ACL
- Using BDI with BEN
- Using Driving Skill
- Manipulate dates
- Manipulate lights
- Using comodel
- Save and restore Simulations
- Using network
- Headless mode
- Using Headless
- Writing Unit Tests
- Ensure model's reproducibility
- Going further with extensions
- Built-in Species
- Built-in Skills
- Built-in Architecture
- Statements
- Data Type
- File Type
- Expressions
- Exhaustive list of GAMA Keywords
- Installing the GIT version
- Developing Extensions
- Introduction to GAMA Java API
- Using GAMA flags
- Creating a release of GAMA
- Documentation generation