scSVAS is an online platform for single cell DNA visualization. It provides versatile utilities for managing, investigating, sharing, and publishing single-cell copy number variation profiles. All visualization are publicly hosted at https://sc.deepomics.org. Offline auxiliary scripts are available at https://github.com/paprikachan/scSVAS.
The recent advance of single-cell copy number variation analysis plays an essential role in addressing intratumor heterogeneity, identifying tumor subgroups, and restoring tumor evolving trajectories at single-cell resolution. Pleasant visualization of copy number analysis results boosts productive scientific exploration, validation, and sharing. Several appealing single-cell copy number figures have been demonstrated in published articles and software. However, those pictures almost lack real-time interaction, and it's hard for researchers to prepare codes to reproduce them from scratch. Moreover, existing single-cell visualization tools are incredibly time-consuming and memory-intensive when it reaches today's large-scale single-cell throughput.
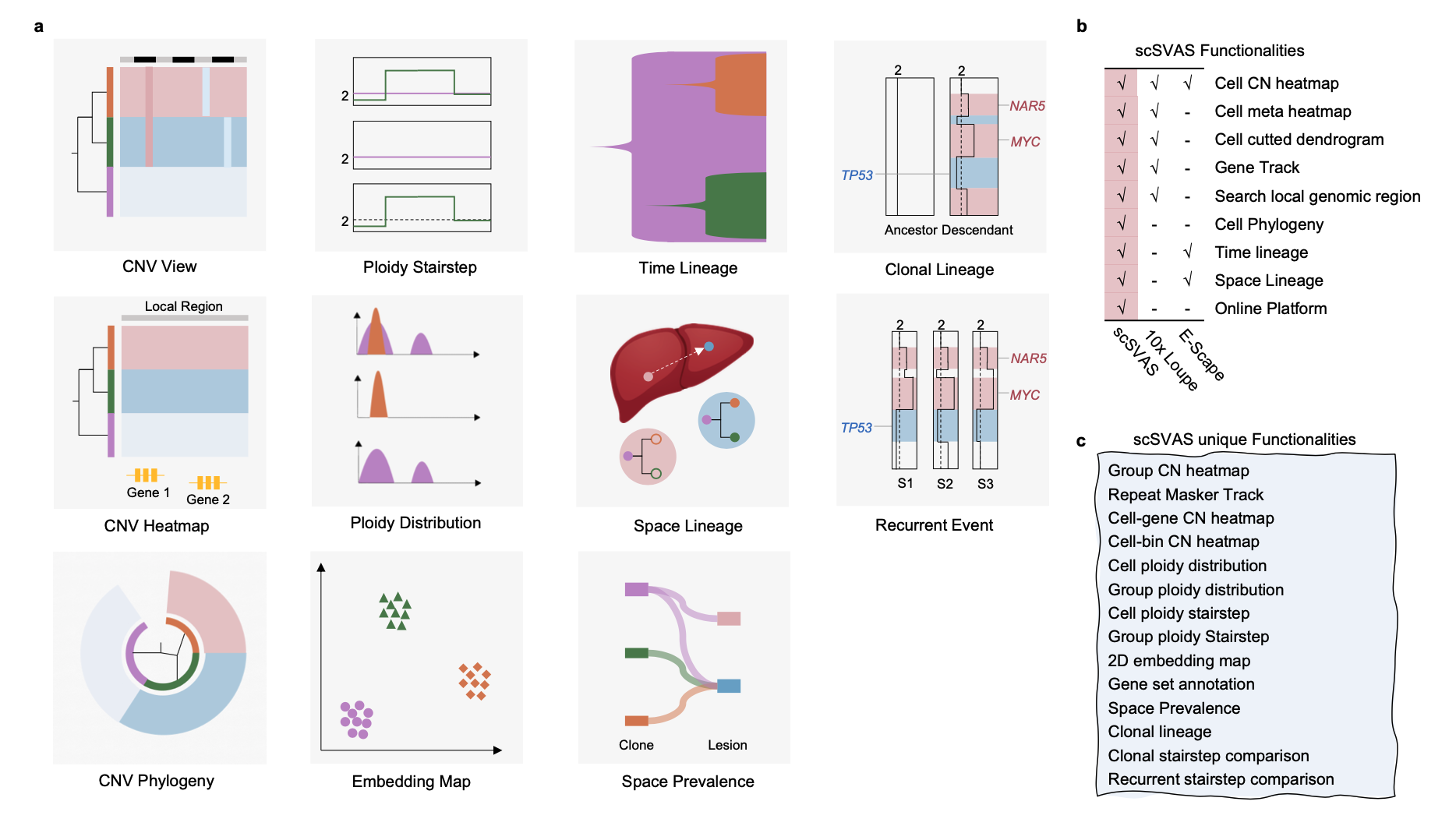 Figure1: overview of scSVAS. The abstract of liver is Designed by macrovector / Freepik.
Figure1: overview of scSVAS. The abstract of liver is Designed by macrovector / Freepik.
We present an online platform scSVAS (https://sc.deepomics.org) for aesthetically-pleasing, real-time interactive, user-friendly single-cell copy number variation analysis, including copy number heatmap view, ploidy stairstep, ploidy distribution, embedding map, time lineage, space lineage, space prevalence, clonal lineage, and recurrent event (Figure1a). scSVAS is specifically designed for large-scale single cells analysis. After uploading the required upstream copy number analysis results, users may make scientific discoveries, and share interactive visualization, and download high-quality publication-ready figures. Compared with other scDNA visualization tools Loupe and E-Scape, scSVAS manifest the most comprehensive functionalities (Figure1b-c).
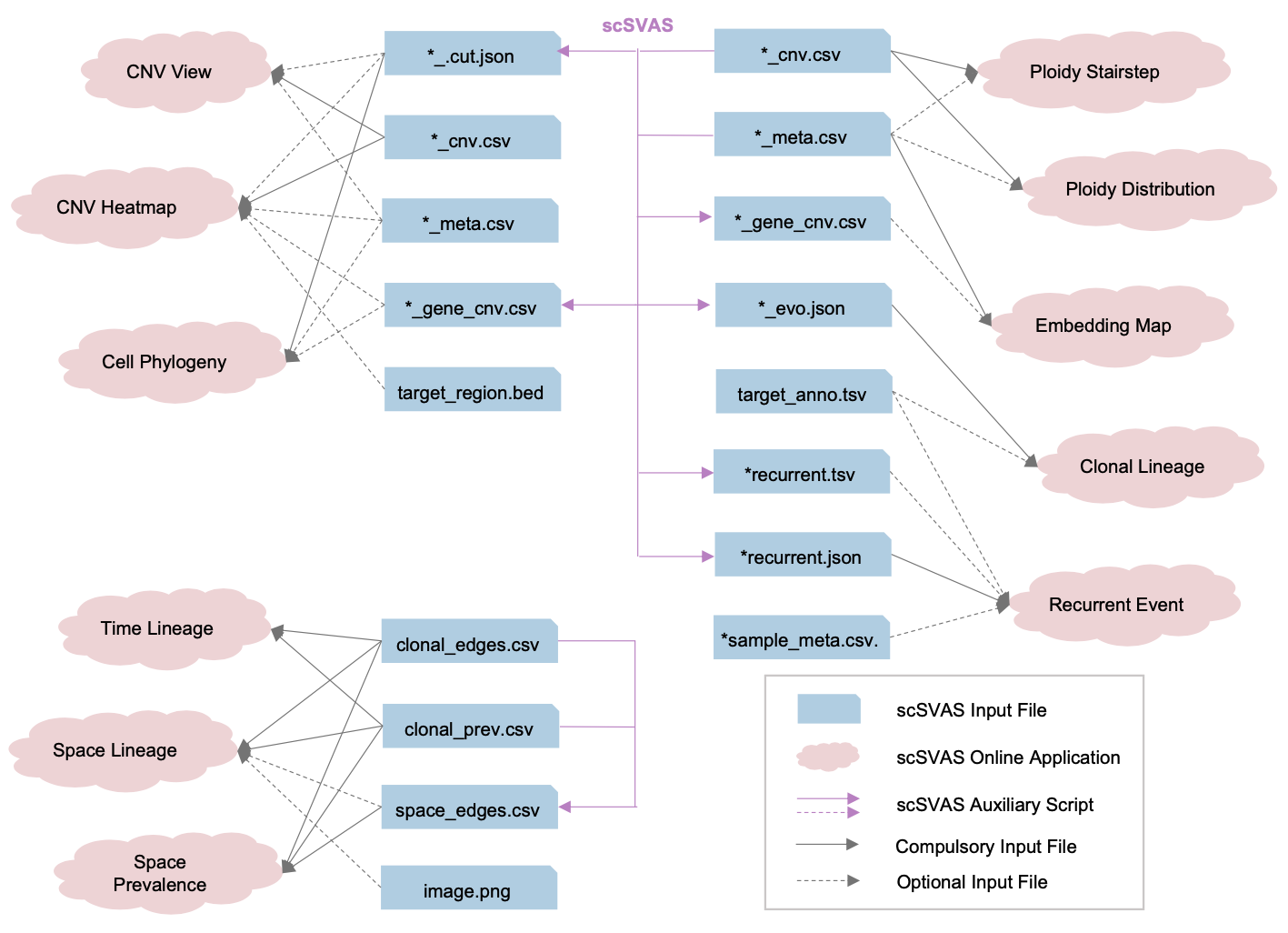 Figure2: the input files required for each scSVAS application
Figure 2 demonstrates the input files required for each scSVAS application. The demo files are available at “Editor” on each application page. The input format are specified in here. The auxiliary scripts are available at
Figure2: the input files required for each scSVAS application
Figure 2 demonstrates the input files required for each scSVAS application. The demo files are available at “Editor” on each application page. The input format are specified in here. The auxiliary scripts are available at ./scripts.
| Firefox | Chrome | Safari | Edge | |
|---|---|---|---|---|
| Linux | √ | √ | - | - |
| Macos | √ | √ | √ | - |
| Windows | √ | √ | - | √ |