This experiment will focus on developing simulations for blood oxygen level-dependent (BOLD) signals in functional Magnetic Resonance Imaging (fMRI) data that serve as the ground truth for Computational Parametric Mapping (CPM), a Bayesian approach for model-based fMRI experiments.
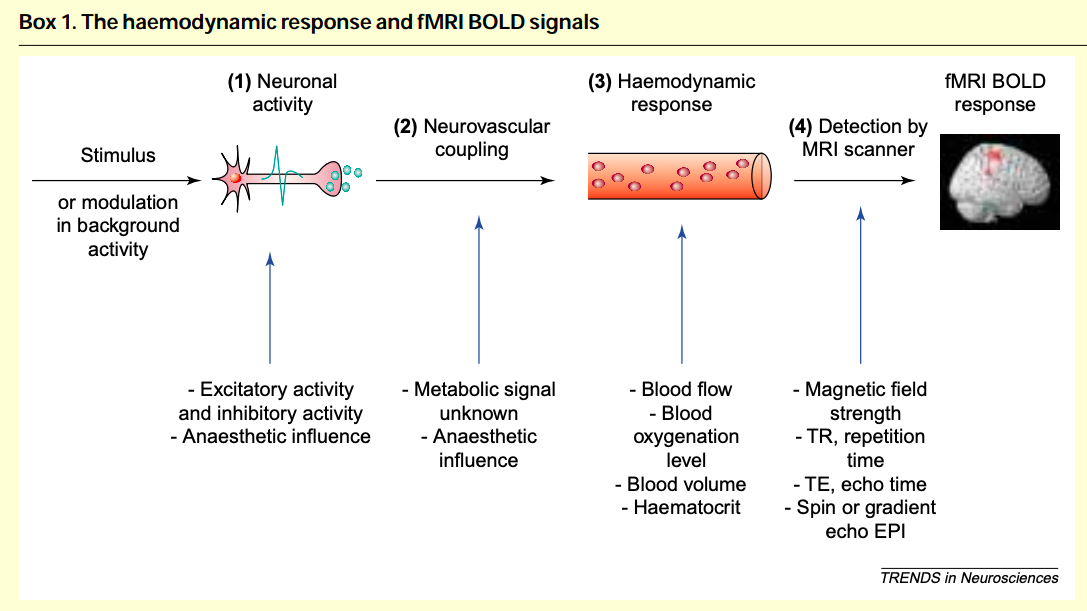
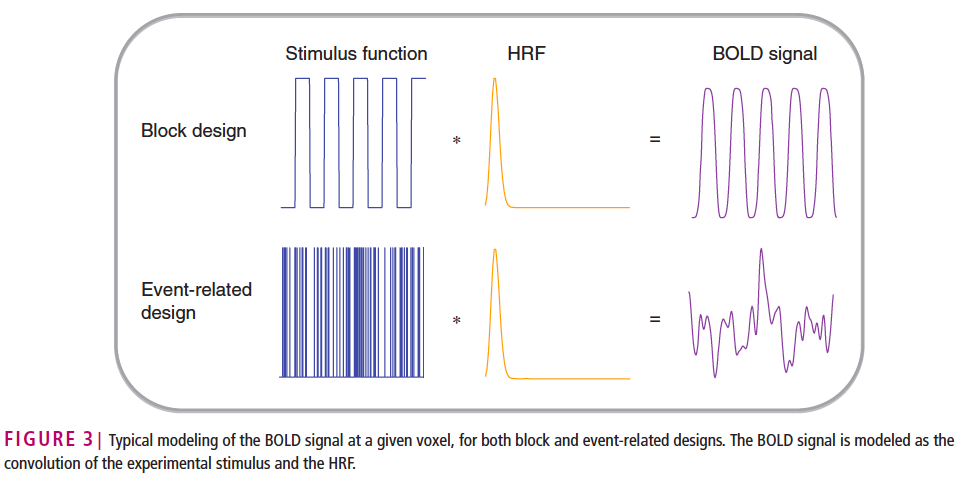
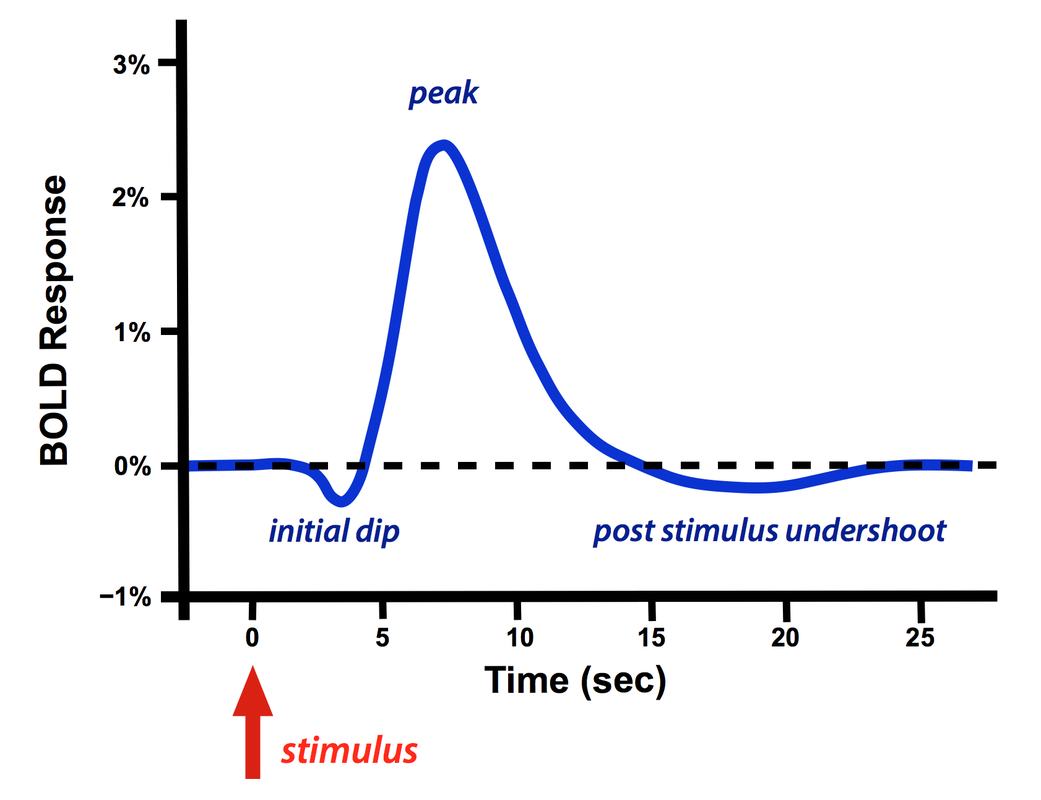
The functional biological process is generated by a stimuli at the finger tip, through many neural pathways and the activation of the hemoglobin response function to manifest as signals in the brain imaging [1]. For functional MRI, there are two experiment designs, as indicated below [2]. A block design gives constant stimulus for a period of time, followed by no stimulus for a while. On the contrary, event related design applies point stimulus, and the resulting BOLD signals over time are voxels in each fMRI image. During the second last step, the BOLD function is generated. It has a initial dip followed by a peak and then a post stimulus undershoot [3]. Given the shape of one single BOLD signal curve, we can approximate it with a inverse gamma distribution. There are two steps for the Bayesian framework: 1. First: Chose a prior. In the current simulation, we chose a conjugate (flat/uninformative) prior for the inverse gamma distribution 2. Second is the power in steven's power law. We want to explore which value of alpha gives the best parameter recovery when designing experiments.
This study is a simplified case with only one beta in the linear model.
For more information, please refer to img/SRP17_Dong_Conference_Post.pdf.
This repository is implemented in MATLAB 3.7, with several dependency functions from [Statistical Parametric Mapping (SPM)] (https://www.fil.ion.ucl.ac.uk/spm/) package developed by UCL for neuroimging.
StarOneBeta.m contains all visualization of the MAP of different power values.
Efficiency.m
Effnojitter.m
While in general, the proposed approach can successfully recover all powers ( value) selected, the neurometric power with
= 1.5 gives the best recovery rate.
[1] Arthurs, O. J., & Boniface, S. (2002). "How well do we understand the neural origins of the fMRI BOLD signal?" Trends in Neurosciences, 25(1), 27–31. [https://doi.org/10.1016/S0166-2236(00)01995-0]
[2] Zhang, L., Guindani, M., & Vannucci, M. (2015). "Bayesian Models for fMRI Data Analysis." Computational Statistics, 7(1), 21–41. [https://doi.org/10.1002/wics.1339]
[3] AD Elster, ELSTER LLC. (2020). "BOLD and Brain Activity". Questions and Answers in MRI. [http://mriquestions.com/does-boldbrain-activity.html]
[4] Relationship between MAP inference and regularization in GLMs. [http://pillowlab.princeton.edu/teaching/statneuro2020/notes/notes13_MAP.pdf]