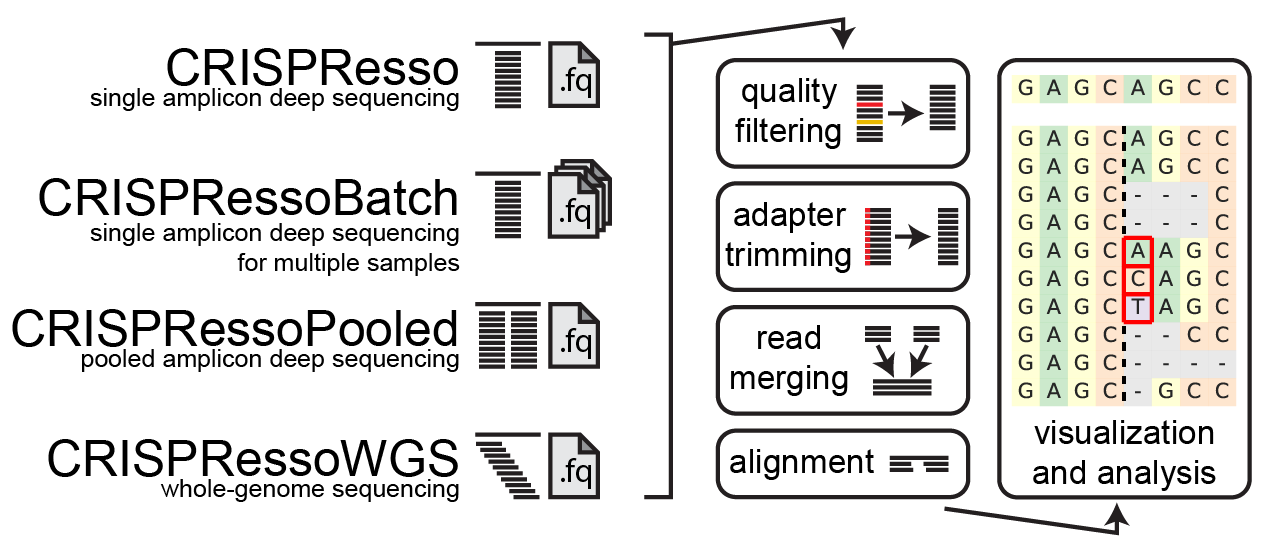
CRISPResso2 is a software pipeline designed to enable rapid and intuitive interpretation of genome editing experiments. A limited web implementation is available at: https://crispresso2.pinellolab.org/.
Briefly, CRISPResso2:
- aligns sequencing reads to a reference sequence
- quantifies insertions, mutations and deletions to determine whether a read is modified or unmodified by genome editing
- summarizes editing results in intuitive plots and datasets
Access the full documentation at https://docs.crispresso.com.
In addition, CRISPResso can be run as part of a larger tool suite:
- CRISPRessoBatch - for analyzing and comparing multiple experimental conditions at the same site
- CRISPRessoPooled - for analyzing multiple amplicons from a pooled amplicon sequencing experiment
- CRISPRessoWGS - for analyzing specific sites in whole-genome sequencing samples
- CRISPRessoCompare - for comparing editing between two samples (e.g., treated vs control)
- CRISPRessoAggregate - for aggregating results from previously-run CRISPResso analyses
CRISPResso2 can be installed in the following ways:
- CRISPResso example runs
- CRISPRessoBatch example runs
- CRISPRessoPooled example runs
- CRISPRessoWGS example runs
- CRISPRessoCompare example runs
- CRISPRessoPooledWGCompare example runs
- CRISPRessoAggregate example runs
If you run into any issues, check out the Troubleshooting page or submit a new discussion.