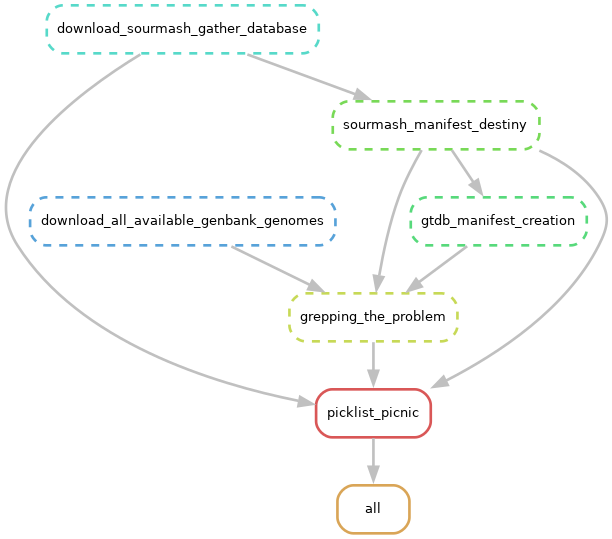
An experimental snakemake workflow repository to remove suppressed or replaced genomes from sourmash workflow.
This workflow uses sourmash's manifest and picklist to exclude supressed and replaced genomes from the prepared GTDB database of sourmash.
To install this workflow:
git clone https://github.com/dib-lab/2023-clean-gtdb.git
or
git clone [email protected]:dib-lab/2023-clean-gtdb.git
cd 2023-clean-gtdb
conda env create --name clean-db --file environment.yml
conda activate clean-db
To run this workflow:
snakemake -s clean-gtdb.snakefile --use-conda --rerun-incomplete -j 1
or
snakemake -s clean-gtdb.snakefile -j 3 --use-conda --rerun-incomplete --resources mem_mb=12000 --cluster "sbatch -t {resources.time_min} -J clean-gtdb -p bmm -n 1 -N 1 -c {threads} --mem={resources.mem_mb}" -k