-
Notifications
You must be signed in to change notification settings - Fork 1
1.6 PD BQ Exercises
These are live data, so the numbers here could change as isolates are added to the system or withdrawn by submitters.
Many SQL tutorials are available on the web, you can easily search them up, and Google Gemini can be useful as well especially when you don't know the names of what you're trying to do. If you need an introduction to BigQuery see https://cloud.google.com/bigquery/docs/introduction-sql See also our 4. Some Useful Links page.
We can count rows very easily using standard SQL. The only thing that is different is the way tables are referred to with "backticks" around the address of the table.
select count(1)
from `ncbi-pathogen-detect.pdbrowser.isolates`
where taxgroup_name = 'Acinetobacter baumannii'You should get a number ~ 36,000
Since not all of the isolates in the Pathogen Detection system have public assemblies in GenBank we can count just the ones that do.
select count(1)
from `ncbi-pathogen-detect.pdbrowser.isolates`
where taxgroup_name = 'Acinetobacter baumannii'
and asm_acc is not NULLYou should get a number ~ 30,000
In order to search repeated record fields like 'AST_phenotypes' we have to use a slightly different technique such as the unnest function which expands the contents of that field into its own table with each row including one row or group of elements from the AST_phenotypes field.
select count(1)
from `ncbi-pathogen-detect.pdbrowser.isolates` i
where taxgroup_name = 'Acinetobacter baumannii'
and asm_acc is not NULL
and (select count(1)
from unnest(i.AST_phenotypes)
where antibiotic like 'tobramycin' ) >=1I got 811 at the time of writing this. A far smaller number than the number of A. baumannii isolates.
Some submitters have run their isolates through multiple phenotyping platforms and have submitted multiple different AST test results for the isolates. How often do different platformst give different results?
The AST browser has data from antibiotic susceptibility tests (AST), including several tests of the drug meropenem. We can search that table and "join" it with itself to identify isolates with conflicting data.
select count(distinct(ast1.biosample_acc))
from `ncbi-pathogen-detect.pdbrowser.ast` ast1
join `ncbi-pathogen-detect.pdbrowser.ast` ast2
on ast1.biosample_acc = ast2.biosample_acc
and ast1.phenotype != ast2.phenotype
and ast1.antibiotic = 'tobramycin'
and ast2.antibiotic = 'tobramycin'148 isolates had multiple tobramycin test results that are different from each other
Very similar to the query above, but this time we want to take a look at the raw data. Looking at the data to see if it makes sense is often a part of good research.
select distinct ast1.target_acc, ast1.biosample_acc, ast1.phenotype,
ast1.mic, ast1.standard, ast1.platform
from `ncbi-pathogen-detect.pdbrowser.ast` ast1
join `ncbi-pathogen-detect.pdbrowser.ast` ast2
on ast1.biosample_acc = ast2.biosample_acc
and ast1.phenotype != ast2.phenotype
and ast1.antibiotic = 'tobramycin'
and ast2.antibiotic = 'tobramycin'
order by ast1.biosample_acc, ast1.mic, ast1.platform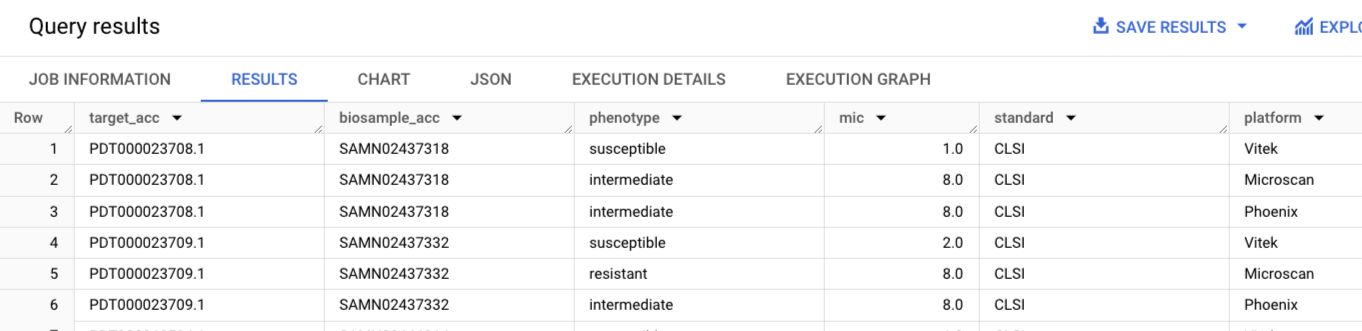
MIC cutoffs can change, and submitters may make mistakes. As of 2023, the CLSI cutoff MIC for tobramycin resistance in Pseudomonas aeruginosa is 8 μg/mL. This means that isolates with a MIC of 8 μg/mL or higher are considered resistant to tobramycin.
What are the MICs for "resistant" isolates?
select mic, count(1), standard
from `ncbi-pathogen-detect.pdbrowser.ast`
where antibiotic = 'tobramycin'
and phenotype = 'resistant'
and taxgroup_name = 'Acinetobacter baumannii'
group by mic, standard
order by micAdding the phenotype to the group by clause and removing resistant phenotype as a requirement allows us to view what people call different MICs.
select mic, phenotype, count(1), standard
from `ncbi-pathogen-detect.pdbrowser.ast`
where antibiotic = 'tobramycin'
and taxgroup_name = 'Acinetobacter baumannii'
group by mic, phenotype, standard
order by mic, phenotypeNotice that at an MIC of 8 there are both intermediate and resistant calls because of changes to the CLSI standard over time.
Above we replicated some of the things that Mike showed you how to do in the browsers. As you have seen above with the joins, SQL has additional analysis capabilities that arent reflected in the browsers. For example we can link between tables to find isolates that are phenotypically resistant to a drug but don't have any resistance elements for that drug.
The AST browser has data from antibiotic susceptibility tests (AST), including several tests of the drug meropenem.
select ast.target_acc, ast.biosample_acc, ast.antibiotic, ast.measurement_sign, ast.mic
from `ncbi-pathogen-detect.pdbrowser.ast` ast
where ast.antibiotic = 'meropenem'
and ast.mic >= 2I got 1,684 rows of meropenem resistance tests at the time of this writing.
9. Find meropenem isolates with MIC >= 2 and no known carbapenem resistance genes or point mutations
SQL joins allow you to merge the data from two tables. We can "join" the AST table data from the above query to the MicroBIGG-E table to identify the isolates that don't have a carbapenem resistance gene or point mutation?
select ast.target_acc, ast.biosample_acc, ast.antibiotic, ast.measurement_sign, ast.mic
from `ncbi-pathogen-detect.pdbrowser.ast` ast
left join `ncbi-pathogen-detect.pdbrowser.microbigge` mb -- all rows from ast and any matching rows from mb
on mb.target_acc = ast.target_acc
and mb.subclass = 'CARBAPENEM'
where ast.antibiotic = 'meropenem'
and ast.mic >= 2
and mb.subclass is NULL -- no rows from mb match the requirements for the join
order by target_accI got 738 resistant tests for isolates without any "CARBAPENEM" resistance elements from MicroBIGG-E.
One complication is that not all isolates have their AMRFinderPlus results in MicroBIGG-E, so the more refined way to do this is to also link to the isolates table to make sure that those isolates have results in MicroBIGG-E (they have public assembly accessions and AMRFinderPlus runs).
select ast.target_acc, ast.biosample_acc, ast.antibiotic, ast.measurement_sign, ast.mic
from `ncbi-pathogen-detect.pdbrowser.ast` ast
left join `ncbi-pathogen-detect.pdbrowser.microbigge` mb -- all rows from ast and any matching rows from mb
on mb.target_acc = ast.target_acc
and mb.subclass = 'CARBAPENEM'
inner join `ncbi-pathogen-detect.pdbrowser.isolates` i
on i.target_acc = ast.target_acc
and i.amrfinderplus_version is not NULL -- AMRFinderPlus was run
and i.asm_acc is not NULL -- There are public assemblies in GenBank
where ast.antibiotic = 'meropenem'
and ast.mic >= 2
and mb.subclass is NULL -- no rows from mb match the requirements for the join
order by target_acc
Now I get 262 isolates with those characteristics.
This work was supported by the National Center for Biotechnology Information of the National Library of Medicine (NLM) and the National Institute of Allergy and Infectious disease (NIAID), National Institutes of Health