diff --git a/annolid/gui/app.py b/annolid/gui/app.py
index daa1bb14..dd62798d 100644
--- a/annolid/gui/app.py
+++ b/annolid/gui/app.py
@@ -315,6 +315,14 @@ def __init__(self,
self.tr("Open video")
)
+ segment_cells = action(
+ self.tr("&Segment Cells"),
+ self._segment_cells,
+ None,
+ "Segment Cells",
+ self.tr("Segment Cells")
+ )
+
advance_params = action(
self.tr("&Advanced Parameters"),
self.set_advanced_params,
@@ -328,6 +336,12 @@ def __init__(self,
)
))
+ segment_cells.setIcon(QtGui.QIcon(
+ str(
+ self.here / "icons/cell_seg.png"
+ )
+ ))
+
open_audio = action(
self.tr("&Open Audio"),
self.openAudio,
@@ -488,6 +502,7 @@ def __init__(self,
_action_tools.append(quality_control)
_action_tools.append(colab)
_action_tools.append(visualization)
+ _action_tools.append(segment_cells)
self.actions.tool = tuple(_action_tools)
self.tools.clear()
@@ -502,6 +517,7 @@ def __init__(self,
utils.addActions(self.menus.file, (models,))
utils.addActions(self.menus.file, (tracks,))
utils.addActions(self.menus.file, (quality_control,))
+ utils.addActions(self.menus.file, (segment_cells,))
utils.addActions(self.menus.file, (downsample_video,))
utils.addActions(self.menus.file, (convert_sleap,))
utils.addActions(self.menus.file, (advance_params,))
@@ -991,6 +1007,46 @@ def _select_sam_model_name(self):
return model_name
+ def _segment_cells(self):
+ if self.filename or len(self.imageList) > 0:
+ from annolid.segmentation.MEDIAR.predict_ensemble import MEDIARPredictor
+ if self.annotation_dir is not None:
+ out_dir_path = self.annotation_dir + '_masks'
+ elif self.filename:
+ out_dir_path = str(Path(self.filename).with_suffix(''))
+ if not os.path.exists(out_dir_path):
+ os.makedirs(out_dir_path, exist_ok=True)
+
+ if self.filename is not None and self.annotation_dir is None:
+ self.annotation_dir = out_dir_path
+ target_link = os.path.join(
+ out_dir_path, os.path.basename(self.filename))
+ if not os.path.islink(target_link):
+ os.symlink(self.filename, target_link)
+ else:
+ logger.info(f"The symlink {target_link} alreay exists.")
+ mediar_predictor = MEDIARPredictor(input_path=self.annotation_dir,
+ output_path=out_dir_path)
+ self.worker = FlexibleWorker(
+ mediar_predictor.conduct_prediction)
+ self.thread = QtCore.QThread()
+ self.worker.moveToThread(self.thread)
+ self.worker.start.connect(self.worker.run)
+ self.worker.finished.connect(self.thread.quit)
+ self.worker.finished.connect(self.worker.deleteLater)
+ self.thread.finished.connect(self.thread.deleteLater)
+ self.worker.finished.connect(lambda:
+ QtWidgets.QMessageBox.about(self,
+ "Cell counting results are ready",
+ f"Please review your results."))
+ self.worker.return_value.connect(
+ lambda shape_list: self.loadShapes(shape_list))
+
+ self.thread.start()
+ self.worker.start.emit()
+ # shape_list = mediar_predictor.conduct_prediction()
+ # self.loadShapes(shape_list)
+
def stop_prediction(self):
# Emit the stop signal to signal the prediction thread to stop
self.pred_worker.stop()
@@ -1009,7 +1065,7 @@ def predict_from_next_frame(self,
if self.pred_worker and self.stop_prediction_flag:
# If prediction is running, stop the prediction
self.stop_prediction()
- elif len(self.canvas.shapes) <= 0:
+ elif len(self.canvas.shapes) <= 0 and self.video_file is not None:
QtWidgets.QMessageBox.about(self,
"No Shapes or Labeled Frames",
f"Please label this frame")
diff --git a/annolid/gui/icons/cell_seg.png b/annolid/gui/icons/cell_seg.png
new file mode 100644
index 00000000..5e116149
Binary files /dev/null and b/annolid/gui/icons/cell_seg.png differ
diff --git a/annolid/segmentation/MEDIAR/.gitignore b/annolid/segmentation/MEDIAR/.gitignore
new file mode 100644
index 00000000..c76d2755
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/.gitignore
@@ -0,0 +1,18 @@
+config/
+*.log
+*.ipynb
+*.ipynb_checkpoints/
+__pycache__/
+results/
+weights/
+wandb/
+data/
+submissions/
+/.vscode
+*.npy
+*.pth
+*.sh
+*.json
+*.out
+*.zip
+*.tiff
diff --git a/annolid/segmentation/MEDIAR/LICENSE b/annolid/segmentation/MEDIAR/LICENSE
new file mode 100644
index 00000000..2161ee79
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/LICENSE
@@ -0,0 +1,21 @@
+MIT License
+
+Copyright (c) 2022 opcrisis
+
+Permission is hereby granted, free of charge, to any person obtaining a copy
+of this software and associated documentation files (the "Software"), to deal
+in the Software without restriction, including without limitation the rights
+to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
+copies of the Software, and to permit persons to whom the Software is
+furnished to do so, subject to the following conditions:
+
+The above copyright notice and this permission notice shall be included in all
+copies or substantial portions of the Software.
+
+THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
+AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
+OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
+SOFTWARE.
diff --git a/annolid/segmentation/MEDIAR/README.md b/annolid/segmentation/MEDIAR/README.md
new file mode 100644
index 00000000..af287217
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/README.md
@@ -0,0 +1,189 @@
+
+# **MEDIAR: Harmony of Data-Centric and Model-Centric for Multi-Modality Microscopy**
+
+
+
+This repository provides an official implementation of [MEDIAR: MEDIAR: Harmony of Data-Centric and Model-Centric for Multi-Modality Microscopy](https://arxiv.org/abs/2212.03465), which achieved the ***"1st winner"*** in the [NeurIPS-2022 Cell Segmentation Challenge](https://neurips22-cellseg.grand-challenge.org/).
+
+To access and try mediar directly, please see links below.
+-  +- [Huggingface Space](https://huggingface.co/spaces/ghlee94/MEDIAR?logs=build)
+- [Napari Plugin](https://github.com/joonkeekim/mediar-napari)
+- [Docker Image](https://hub.docker.com/repository/docker/joonkeekim/mediar/general)
+
+# 1. MEDIAR Overview
+
+- [Huggingface Space](https://huggingface.co/spaces/ghlee94/MEDIAR?logs=build)
+- [Napari Plugin](https://github.com/joonkeekim/mediar-napari)
+- [Docker Image](https://hub.docker.com/repository/docker/joonkeekim/mediar/general)
+
+# 1. MEDIAR Overview
+ +
+MEIDAR is a framework for efficient cell instance segmentation of multi-modality microscopy images. The above figure illustrates an overview of our approach. MEDIAR harmonizes data-centric and model-centric approaches as the learning and inference strategies, achieving a **0.9067** Mean F1-score on the validation datasets. We provide a brief description of methods that combined in the MEDIAR. Please refer to our paper for more information.
+# 2. Methods
+
+## **Data-Centric**
+- **Cell Aware Augmentation** : We apply two novel cell-aware augmentations. *cell-wisely intensity is randomization* (Cell Intensity Diversification) and *cell-wise boundary pixels exclusion* in the label. The boundary exclusion is adopted only in the pre-training phase.
+
+- **Two-phase Pretraining and Fine-tuning** : To extract knowledge from large public datasets, we first pretrained our model on public sets, then fine-tune.
+ - Pretraining : We use 7,2412 labeled images from four public datasets for pretraining: OmniPose, CellPose, LiveCell and DataScienceBowl-2018. MEDIAR takes two different phases for the pretraining. the MEDIAR-Former model with encoder parameters initialized from ImageNet-1k pretraining.
+
+ - Fine-tuning : We use two different model for ensemble. First model is fine-tuned 200 epochs using target datasets. Second model is fine-tuned 25 epochs using both target and public datsets.
+
+- **Modality Discovery & Amplified Sampling** : To balance towards the latent modalities in the datasets, we conduct K-means clustering and discover 40 modalities. In the training phase, we over-sample the minor cluster samples.
+
+- **Cell Memory Replay** : We concatenate the data from the public dataset with a small portion to the batch and train with boundary-excluded labels.
+
+## **Model-Centric**
+- **MEDIAR-Former Architecture** : MEDIAR-Former follows the design paradigm of U-Net, but use SegFormer and MA-Net for the encoder and decoder. The two heads of MEDIAR-Former predicts cell probability and gradieng flow.
+
+
+
+MEIDAR is a framework for efficient cell instance segmentation of multi-modality microscopy images. The above figure illustrates an overview of our approach. MEDIAR harmonizes data-centric and model-centric approaches as the learning and inference strategies, achieving a **0.9067** Mean F1-score on the validation datasets. We provide a brief description of methods that combined in the MEDIAR. Please refer to our paper for more information.
+# 2. Methods
+
+## **Data-Centric**
+- **Cell Aware Augmentation** : We apply two novel cell-aware augmentations. *cell-wisely intensity is randomization* (Cell Intensity Diversification) and *cell-wise boundary pixels exclusion* in the label. The boundary exclusion is adopted only in the pre-training phase.
+
+- **Two-phase Pretraining and Fine-tuning** : To extract knowledge from large public datasets, we first pretrained our model on public sets, then fine-tune.
+ - Pretraining : We use 7,2412 labeled images from four public datasets for pretraining: OmniPose, CellPose, LiveCell and DataScienceBowl-2018. MEDIAR takes two different phases for the pretraining. the MEDIAR-Former model with encoder parameters initialized from ImageNet-1k pretraining.
+
+ - Fine-tuning : We use two different model for ensemble. First model is fine-tuned 200 epochs using target datasets. Second model is fine-tuned 25 epochs using both target and public datsets.
+
+- **Modality Discovery & Amplified Sampling** : To balance towards the latent modalities in the datasets, we conduct K-means clustering and discover 40 modalities. In the training phase, we over-sample the minor cluster samples.
+
+- **Cell Memory Replay** : We concatenate the data from the public dataset with a small portion to the batch and train with boundary-excluded labels.
+
+## **Model-Centric**
+- **MEDIAR-Former Architecture** : MEDIAR-Former follows the design paradigm of U-Net, but use SegFormer and MA-Net for the encoder and decoder. The two heads of MEDIAR-Former predicts cell probability and gradieng flow.
+
+ +
+- **Gradient Flow Tracking** : We utilize gradient flow tracking proposed by [CellPose](https://github.com/MouseLand/cellpose).
+
+- **Ensemble with Stochastic TTA**: During the inference, the MEIDAR conduct prediction as sliding-window manner with importance map generated by the gaussian filter. We use two fine-tuned models from phase1 and phase2 pretraining, and ensemble their outputs by summation. For each outputs, test-time augmentation is used.
+# 3. Experiments
+
+### **Dataset**
+- Official Dataset
+ - We are provided the target dataset from [Weakly Supervised Cell Segmentation in Multi-modality High-Resolution Microscopy Images](https://neurips22-cellseg.grand-challenge.org/). It consists of 1,000 labeled images, 1,712 unlabeled images and 13 unlabeled whole slide image from various microscopy types, tissue types, and staining types. Validation set is given with 101 images including 1 whole slide image.
+
+- Public Dataset
+ - [OmniPose](http://www.cellpose.org/dataset_omnipose) : contains mixtures of 14 bacterial species. We only use 611 bacterial cell microscopy images and discard 118 worm images.
+ - [CellPose](https://www.cellpose.org/dataset) : includes Cytoplasm, cellular microscopy, fluorescent cells images. We used 551 images by discarding 58 non-microscopy images. We convert all images as gray-scale.
+ - [LiveCell](https://github.com/sartorius-research/LIVECell) : is a large-scale dataset with 5,239 images containing 1,686,352 individual cells annotated by trained crowdsources from 8 distinct cell types.
+ - [DataScienceBowl 2018](https://www.kaggle.com/competitions/sartorius-cell-instance-segmentation/overview) : 841 images contain 37,333 cells from 22 cell types, 15 image resolutions, and five visually similar groups.
+
+### **Testing steps**
+- **Ensemble Prediction with TTA** : MEDIAR uses sliding-window inference with the overlap size between the adjacent patches as 0.6 and gaussian importance map. To predict the different views on the image, MEDIAR uses Test-Time Augmentation (TTA) for the model prediction and ensemble two models described in **Two-phase Pretraining and Fine-tuning**.
+
+- **Inference time** : MEDIAR conducts most images in less than 1sec and it depends on the image size and the number of cells, even with ensemble prediction with TTA. Detailed evaluation-time results are in the paper.
+
+### **Preprocessing & Augmentations**
+| Strategy | Type | Probability |
+|----------|:-------------|------|
+| `Clip` | Pre-processing | . |
+| `Normalization` | Pre-processing | . |
+| `Scale Intensity` | Pre-processing | . |
+| `Zoom` | Spatial Augmentation | 0.5 |
+| `Spatial Crop` | Spatial Augmentation | 1.0 |
+| `Axis Flip` | Spatial Augmentation | 0.5 |
+| `Rotation` | Spatial Augmentation | 0.5 |
+| `Cell-Aware Intensity` | Intensity Augmentation | 0.25 |
+| `Gaussian Noise` | Intensity Augmentation | 0.25 |
+| `Contrast Adjustment` | Intensity Augmentation | 0.25 |
+| `Gaussian Smoothing` | Intensity Augmentation | 0.25 |
+| `Histogram Shift` | Intensity Augmentation | 0.25 |
+| `Gaussian Sharpening` | Intensity Augmentation | 0.25 |
+| `Boundary Exclusion` | Others | . |
+
+
+| Learning Setups | Pretraining | Fine-tuning |
+|----------------------------------------------------------------------|---------------------------------------------------------|---------------------------------------------------------|
+| Initialization (Encoder) | Imagenet-1k pretrained | from Pretraining |
+| Initialization (Decoder, Head) | He normal initialization | from Pretraining|
+| Batch size | 9 | 9 |
+| Total epochs | 80 (60) | 200 (25) |
+| Optimizer | AdamW | AdamW |
+| Initial learning rate (lr) | 5e-5 | 2e-5 |
+| Lr decay schedule | Cosine scheduler (100 interval) | Cosine scheduler (100 interval) |
+| Loss function | MSE, BCE | MSE, BCE |
+
+# 4. Results
+### **Validation Dataset**
+- Quantitative Evaluation
+ - Our MEDIAR achieved **0.9067** validation mean F1-score.
+- Qualitative Evaluation
+
+
+- **Gradient Flow Tracking** : We utilize gradient flow tracking proposed by [CellPose](https://github.com/MouseLand/cellpose).
+
+- **Ensemble with Stochastic TTA**: During the inference, the MEIDAR conduct prediction as sliding-window manner with importance map generated by the gaussian filter. We use two fine-tuned models from phase1 and phase2 pretraining, and ensemble their outputs by summation. For each outputs, test-time augmentation is used.
+# 3. Experiments
+
+### **Dataset**
+- Official Dataset
+ - We are provided the target dataset from [Weakly Supervised Cell Segmentation in Multi-modality High-Resolution Microscopy Images](https://neurips22-cellseg.grand-challenge.org/). It consists of 1,000 labeled images, 1,712 unlabeled images and 13 unlabeled whole slide image from various microscopy types, tissue types, and staining types. Validation set is given with 101 images including 1 whole slide image.
+
+- Public Dataset
+ - [OmniPose](http://www.cellpose.org/dataset_omnipose) : contains mixtures of 14 bacterial species. We only use 611 bacterial cell microscopy images and discard 118 worm images.
+ - [CellPose](https://www.cellpose.org/dataset) : includes Cytoplasm, cellular microscopy, fluorescent cells images. We used 551 images by discarding 58 non-microscopy images. We convert all images as gray-scale.
+ - [LiveCell](https://github.com/sartorius-research/LIVECell) : is a large-scale dataset with 5,239 images containing 1,686,352 individual cells annotated by trained crowdsources from 8 distinct cell types.
+ - [DataScienceBowl 2018](https://www.kaggle.com/competitions/sartorius-cell-instance-segmentation/overview) : 841 images contain 37,333 cells from 22 cell types, 15 image resolutions, and five visually similar groups.
+
+### **Testing steps**
+- **Ensemble Prediction with TTA** : MEDIAR uses sliding-window inference with the overlap size between the adjacent patches as 0.6 and gaussian importance map. To predict the different views on the image, MEDIAR uses Test-Time Augmentation (TTA) for the model prediction and ensemble two models described in **Two-phase Pretraining and Fine-tuning**.
+
+- **Inference time** : MEDIAR conducts most images in less than 1sec and it depends on the image size and the number of cells, even with ensemble prediction with TTA. Detailed evaluation-time results are in the paper.
+
+### **Preprocessing & Augmentations**
+| Strategy | Type | Probability |
+|----------|:-------------|------|
+| `Clip` | Pre-processing | . |
+| `Normalization` | Pre-processing | . |
+| `Scale Intensity` | Pre-processing | . |
+| `Zoom` | Spatial Augmentation | 0.5 |
+| `Spatial Crop` | Spatial Augmentation | 1.0 |
+| `Axis Flip` | Spatial Augmentation | 0.5 |
+| `Rotation` | Spatial Augmentation | 0.5 |
+| `Cell-Aware Intensity` | Intensity Augmentation | 0.25 |
+| `Gaussian Noise` | Intensity Augmentation | 0.25 |
+| `Contrast Adjustment` | Intensity Augmentation | 0.25 |
+| `Gaussian Smoothing` | Intensity Augmentation | 0.25 |
+| `Histogram Shift` | Intensity Augmentation | 0.25 |
+| `Gaussian Sharpening` | Intensity Augmentation | 0.25 |
+| `Boundary Exclusion` | Others | . |
+
+
+| Learning Setups | Pretraining | Fine-tuning |
+|----------------------------------------------------------------------|---------------------------------------------------------|---------------------------------------------------------|
+| Initialization (Encoder) | Imagenet-1k pretrained | from Pretraining |
+| Initialization (Decoder, Head) | He normal initialization | from Pretraining|
+| Batch size | 9 | 9 |
+| Total epochs | 80 (60) | 200 (25) |
+| Optimizer | AdamW | AdamW |
+| Initial learning rate (lr) | 5e-5 | 2e-5 |
+| Lr decay schedule | Cosine scheduler (100 interval) | Cosine scheduler (100 interval) |
+| Loss function | MSE, BCE | MSE, BCE |
+
+# 4. Results
+### **Validation Dataset**
+- Quantitative Evaluation
+ - Our MEDIAR achieved **0.9067** validation mean F1-score.
+- Qualitative Evaluation
+ +
+- Failure Cases
+
+
+- Failure Cases
+ +
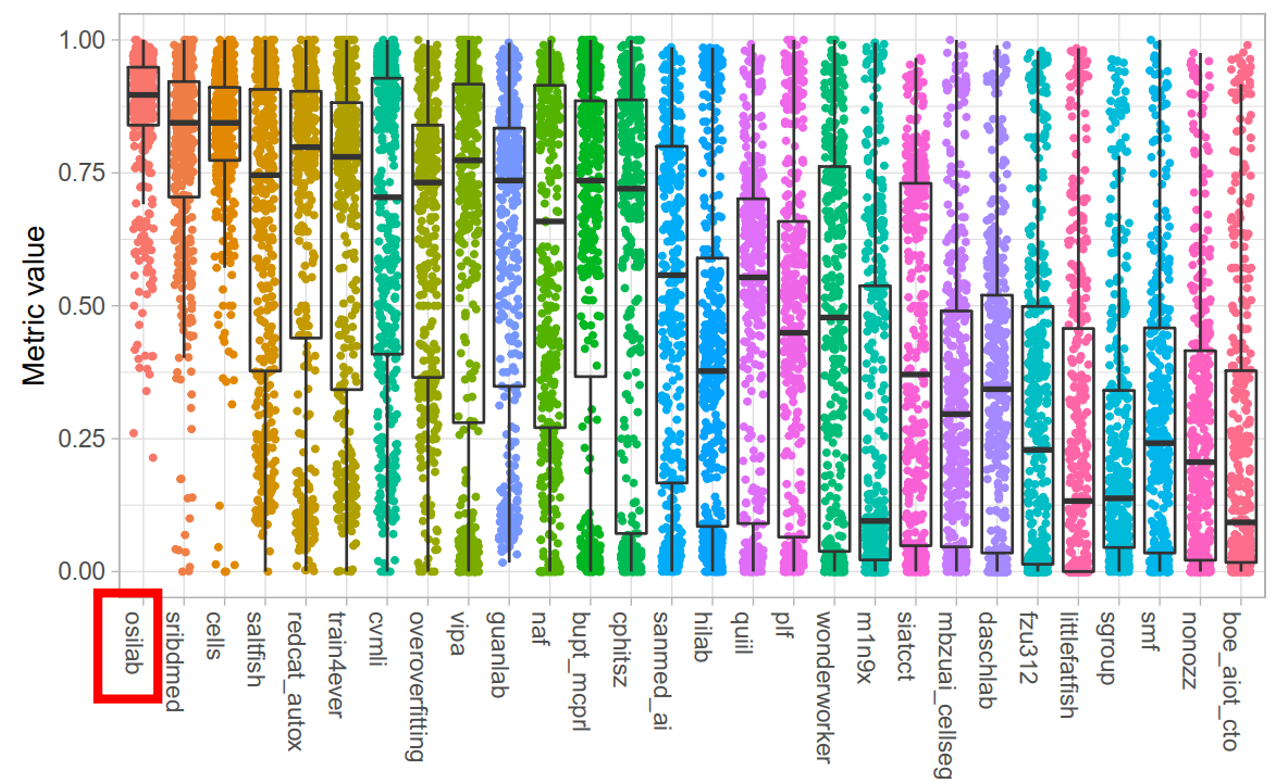
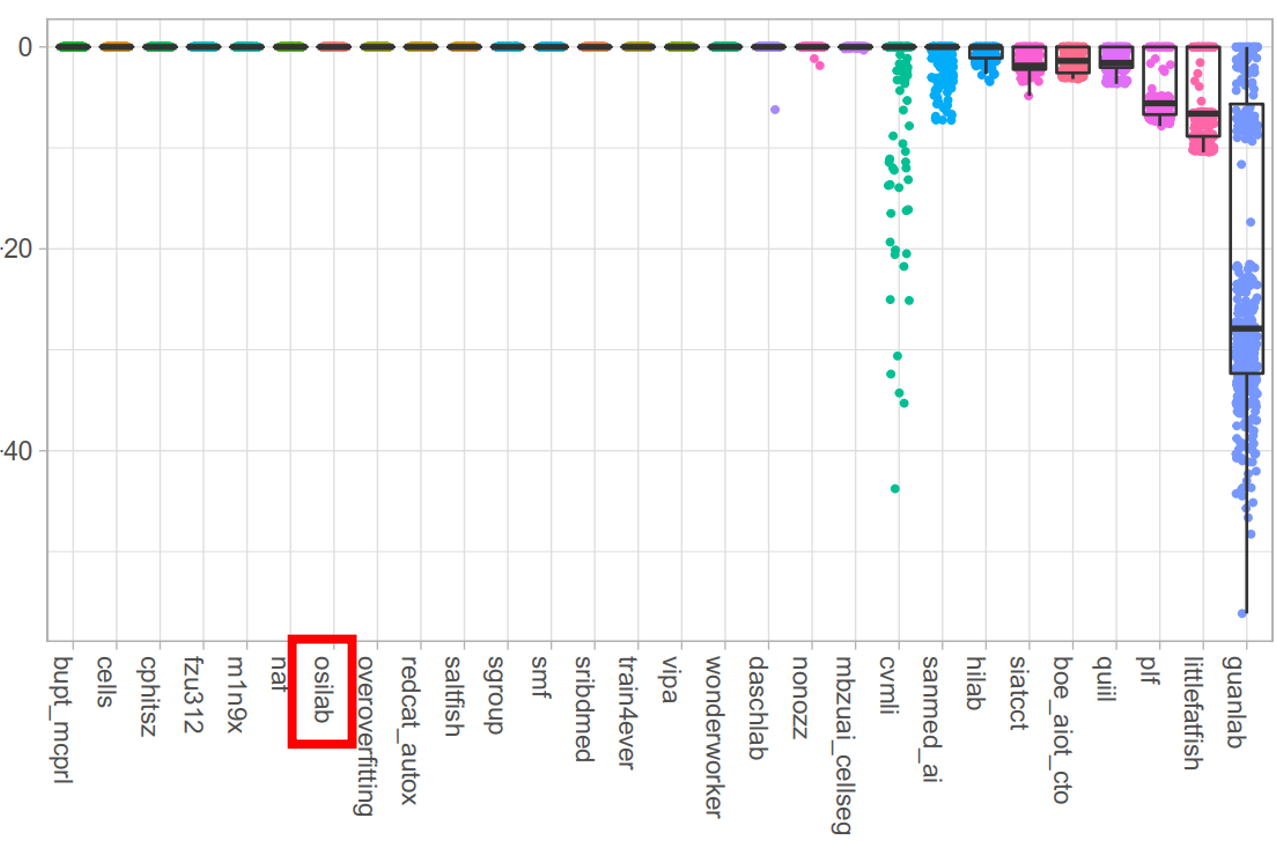
+### **Test Dataset**
+
+
+
+
+# 5. Reproducing
+
+### **Our Environment**
+| Computing Infrastructure| |
+|-------------------------|----------------------------------------------------------------------|
+| System | Ubuntu 18.04.5 LTS |
+| CPU | AMD EPYC 7543 32-Core Processor CPU@2.26GHz |
+| RAM | 500GB; 3.125MT/s |
+| GPU (number and type) | NVIDIA A5000 (24GB) 2ea |
+| CUDA version | 11.7 |
+| Programming language | Python 3.9 |
+| Deep learning framework | Pytorch (v1.12, with torchvision v0.13.1) |
+| Code dependencies | MONAI (v0.9.0), Segmentation Models (v0.3.0) |
+| Specific dependencies | None |
+
+To install requirements:
+
+```
+pip install -r requirements.txt
+wandb off
+```
+
+## Dataset
+- The datasets directories under the root should the following structure:
+
+```
+ Root
+ ├── Datasets
+ │ ├── images (images can have various extensions: .tif, .tiff, .png, .bmp ...)
+ │ │ ├── cell_00001.png
+ │ │ ├── cell_00002.tif
+ │ │ ├── cell_00003.xxx
+ │ │ ├── ...
+ │ └── labels (labels must have .tiff extension.)
+ │ │ ├── cell_00001_label.tiff
+ │ │ ├── cell_00002.label.tiff
+ │ │ ├── cell_00003.label.tiff
+ │ │ ├── ...
+ └── ...
+```
+
+Before execute the codes, run the follwing code to generate path mappting json file:
+
+```python
+python ./generate_mapping.py --root=
+```
+
+## Training
+
+To train the model(s) in the paper, run the following command:
+
+```python
+python ./main.py --config_path=
+```
+Configuration files are in `./config/*`. We provide the pretraining, fine-tuning, and prediction configs. You can refer to the configuration options in the `./config/mediar_example.json`. We also implemented the official challenge baseline code in our framework. You can run the baseline code by running the `./config/baseline.json`.
+
+## Inference
+
+To conduct prediction on the testing cases, run the following command:
+
+```python
+python predict.py --config_path=
+```
+
+## Evaluation
+If you have the labels run the following command for evaluation:
+
+```python
+python ./evaluate.py --pred_path= --gt_path=
+```
+
+The configuration files for `predict.py` is slightly different. Please refer to the config files in `./config/step3_prediction/*`.
+## Trained Models
+
+You can download MEDIAR pretrained and finetuned models here:
+
+- [Google Drive Link](https://drive.google.com/drive/folders/1RgMxHIT7WsKNjir3wXSl7BrzlpS05S18?usp=share_link).
+
+## Citation of this Work
+```
+@article{lee2022mediar,
+ title={Mediar: Harmony of data-centric and model-centric for multi-modality microscopy},
+ author={Lee, Gihun and Kim, SangMook and Kim, Joonkee and Yun, Se-Young},
+ journal={arXiv preprint arXiv:2212.03465},
+ year={2022}
+}
+```
+
diff --git a/annolid/segmentation/MEDIAR/SetupDict.py b/annolid/segmentation/MEDIAR/SetupDict.py
new file mode 100644
index 00000000..d9b7aebd
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/SetupDict.py
@@ -0,0 +1,39 @@
+import torch.optim as optim
+import torch.optim.lr_scheduler as lr_scheduler
+import monai
+
+from annolid.segmentation.MEDIAR import core
+from annolid.segmentation.MEDIAR.train_tools import models
+from annolid.segmentation.MEDIAR.train_tools.models import *
+
+__all__ = ["TRAINER", "OPTIMIZER", "SCHEDULER"]
+
+TRAINER = {
+ "baseline": core.Baseline.Trainer,
+ "mediar": core.MEDIAR.Trainer,
+}
+
+PREDICTOR = {
+ "baseline": core.Baseline.Predictor,
+ "mediar": core.MEDIAR.Predictor,
+ "ensemble_mediar": core.MEDIAR.EnsemblePredictor,
+}
+
+MODELS = {
+ "unet": monai.networks.nets.UNet,
+ "unetr": monai.networks.nets.unetr.UNETR,
+ "swinunetr": monai.networks.nets.SwinUNETR,
+ "mediar-former": models.MEDIARFormer,
+}
+
+OPTIMIZER = {
+ "sgd": optim.SGD,
+ "adam": optim.Adam,
+ "adamw": optim.AdamW,
+}
+
+SCHEDULER = {
+ "step": lr_scheduler.StepLR,
+ "multistep": lr_scheduler.MultiStepLR,
+ "cosine": lr_scheduler.CosineAnnealingLR,
+}
diff --git a/annolid/segmentation/MEDIAR/__init__.py b/annolid/segmentation/MEDIAR/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/annolid/segmentation/MEDIAR/config/baseline.json b/annolid/segmentation/MEDIAR/config/baseline.json
new file mode 100644
index 00000000..186910a4
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/baseline.json
@@ -0,0 +1,60 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/data/CellSeg/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "tuning_mapping_file": "/home/gihun/CellSeg/train_tools/data_utils/mapping_tuning.json",
+ "batch_size": 8,
+ "valid_portion": 0.1
+ },
+ "unlabeled":{
+ "enabled": false
+ },
+ "public":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "swinunetr",
+ "params": {
+ "img_size": 512,
+ "in_channels": 3,
+ "out_channels": 3,
+ "spatial_dims": 2
+ },
+ "pretrained":{
+ "enabled": false
+ }
+ },
+ "trainer": {
+ "name": "baseline",
+ "params": {
+ "num_epochs": 200,
+ "valid_frequency": 1,
+ "device": "cuda:0",
+ "algo_params": {}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "params": {"lr": 5e-5}
+ },
+ "scheduler":{
+ "enabled": false
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/data/CellSeg/Official/TuningSet",
+ "output_path": "./results/baseline",
+ "make_submission": true,
+ "exp_name": "baseline",
+ "algo_params": {}
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Baseline",
+ "name": "baseline"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/mediar_example.json b/annolid/segmentation/MEDIAR/config/mediar_example.json
new file mode 100644
index 00000000..11c99495
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/mediar_example.json
@@ -0,0 +1,72 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/data/CellSeg/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "amplified": false,
+ "batch_size": 8,
+ "valid_portion": 0.1
+ },
+ "public":{
+ "enabled": true,
+ "params":{
+ "root": "/home/gihun/data/CellSeg/",
+ "mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "batch_size": 1
+ }
+ },
+ "unlabeled":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {
+ "encoder_name": "mit_b5",
+ "encoder_weights": "imagenet",
+ "decoder_channels": [1024, 512, 256, 128, 64],
+ "decoder_pab_channels": 256,
+ "in_channels": 3,
+ "classes": 3
+ },
+ "pretrained":{
+ "enabled": false,
+ "weights": "./weights/pretrained/phase2.pth",
+ "strict": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 200,
+ "valid_frequency": 1,
+ "device": "cuda:0",
+ "amp": true,
+ "algo_params": {"with_public": false}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "params": {"lr": 5e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 100, "eta_min": 1e-7}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/data/CellSeg/Official/TuningSet",
+ "output_path": "./mediar_example",
+ "make_submission": true,
+ "exp_name": "mediar_example",
+ "algo_params": {"use_tta": false}
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "MEDIAR",
+ "name": "mediar_example"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/pred/pred_mediar.json b/annolid/segmentation/MEDIAR/config/pred/pred_mediar.json
new file mode 100644
index 00000000..65493771
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/pred/pred_mediar.json
@@ -0,0 +1,17 @@
+{
+ "pred_setups":{
+ "name": "medair",
+ "input_path":"input_path",
+ "output_path": "./test",
+ "make_submission": true,
+ "model_path": "model_path",
+ "device": "cuda:0",
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": false
+ }
+ }
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step1_pretraining/phase1.json b/annolid/segmentation/MEDIAR/config/step1_pretraining/phase1.json
new file mode 100644
index 00000000..a8baee70
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step1_pretraining/phase1.json
@@ -0,0 +1,55 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "mapping_file_tuning": "/home/gihun/MEDIAR/train_tools/data_utils/mapping_tuning.json",
+ "batch_size": 9,
+ "valid_portion": 0
+ },
+ "public":{
+ "enabled": false,
+ "params":{}
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 80,
+ "valid_frequency": 10,
+ "device": "cuda:0",
+ "amp": true,
+ "algo_params": {"with_public": false}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "ft_rate": 1.0,
+ "params": {"lr": 5e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 80, "eta_min": 1e-6}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./mediar_pretrained_phase1",
+ "make_submission": false
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Pretraining",
+ "name": "phase1"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step1_pretraining/phase2.json b/annolid/segmentation/MEDIAR/config/step1_pretraining/phase2.json
new file mode 100644
index 00000000..b13e4bb4
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step1_pretraining/phase2.json
@@ -0,0 +1,58 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "mapping_file_tuning": "/home/gihun/MEDIAR/train_tools/data_utils/mapping_tuning.json",
+ "join_mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "batch_size": 9,
+ "valid_portion": 0
+ },
+ "unlabeled":{
+ "enabled": false
+ },
+ "public":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 60,
+ "valid_frequency": 10,
+ "device": "cuda:0",
+ "amp": true,
+ "algo_params": {"with_public": false}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "ft_rate": 1.0,
+ "params": {"lr": 5e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 60, "eta_min": 1e-6}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./mediar_pretrain_phase2",
+ "make_submission": false
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Pretraining",
+ "name": "phase2"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning1.json b/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning1.json
new file mode 100644
index 00000000..dbdea432
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning1.json
@@ -0,0 +1,66 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "mapping_file_tuning": "/home/gihun/MEDIAR/train_tools/data_utils/mapping_tuning.json",
+ "amplified": true,
+ "batch_size": 8,
+ "valid_portion": 0.0
+ },
+ "public":{
+ "enabled": false,
+ "params":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "batch_size": 1
+ }
+ },
+ "unlabeled":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": true,
+ "weights": "./weights/pretrained/phase1.pth",
+ "strict": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 200,
+ "valid_frequency": 1,
+ "device": "cuda:7",
+ "amp": true,
+ "algo_params": {"with_public": false}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "params": {"lr": 2e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 100, "eta_min": 1e-7}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./results/",
+ "make_submission": true,
+ "exp_name": "mediar_from_phase1",
+ "algo_params": {"use_tta": false}
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Fine-tuning",
+ "name": "from_phase1"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning2.json b/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning2.json
new file mode 100644
index 00000000..149a0f5d
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning2.json
@@ -0,0 +1,66 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "mapping_file_tuning": "/home/gihun/MEDIAR/train_tools/data_utils/mapping_tuning.json",
+ "amplified": true,
+ "batch_size": 8,
+ "valid_portion": 0.0
+ },
+ "public":{
+ "enabled": true,
+ "params":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "batch_size": 1
+ }
+ },
+ "unlabeled":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": true,
+ "weights": "./weights/pretrained/phase2.pth",
+ "strict": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 50,
+ "valid_frequency": 1,
+ "device": "cuda:0",
+ "amp": true,
+ "algo_params": {"with_public": true}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "params": {"lr": 2e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 100, "eta_min": 1e-7}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./results/from_phase2",
+ "make_submission": true,
+ "exp_name": "mediar_from_phase2",
+ "algo_params": {"use_tta": false}
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Fine-tuning",
+ "name": "from_phase2"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step3_prediction/base_prediction.json b/annolid/segmentation/MEDIAR/config/step3_prediction/base_prediction.json
new file mode 100644
index 00000000..e253daea
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step3_prediction/base_prediction.json
@@ -0,0 +1,16 @@
+{
+ "pred_setups":{
+ "name": "mediar",
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./results/mediar_base_prediction",
+ "make_submission": true,
+ "model_path": "./weights/finetuned/from_phase1.pth",
+ "device": "cuda:7",
+ "model":{
+ "name": "mediar-former",
+ "params": {}
+ },
+ "exp_name": "mediar_p1_base",
+ "algo_params": {"use_tta": false}
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step3_prediction/ensemble_tta.json b/annolid/segmentation/MEDIAR/config/step3_prediction/ensemble_tta.json
new file mode 100644
index 00000000..4d3910b6
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step3_prediction/ensemble_tta.json
@@ -0,0 +1,23 @@
+{
+ "pred_setups":{
+ "name": "ensemble_mediar",
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./results/mediar_ensemble_tta",
+ "make_submission": true,
+ "model_path1": "./weights/finetuned/from_phase1.pth",
+ "model_path2": "./weights/finetuned/from_phase2.pth",
+ "device": "cuda:0",
+ "model":{
+ "name": "mediar-former",
+ "params": {
+ "encoder_name":"mit_b5",
+ "decoder_channels": [1024, 512, 256, 128, 64],
+ "decoder_pab_channels": 256,
+ "in_channels":3,
+ "classes":3
+ }
+ },
+ "exp_name": "mediar_ensemble_tta",
+ "algo_params": {"use_tta": true}
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/core/BasePredictor.py b/annolid/segmentation/MEDIAR/core/BasePredictor.py
new file mode 100644
index 00000000..c1c9753d
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/BasePredictor.py
@@ -0,0 +1,126 @@
+import torch
+import numpy as np
+import time
+import os
+import tifffile as tif
+
+from datetime import datetime
+from zipfile import ZipFile
+from pytz import timezone
+from annolid.segmentation.MEDIAR.train_tools.data_utils.transforms import get_pred_transforms
+
+
+class BasePredictor:
+ def __init__(
+ self,
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission=False,
+ exp_name=None,
+ algo_params=None,
+ ):
+ self.model = model
+ self.device = device
+ self.input_path = input_path
+ self.output_path = output_path
+ self.make_submission = make_submission
+ self.exp_name = exp_name
+
+ # Assign algoritm-specific arguments
+ if algo_params:
+ self.__dict__.update((k, v) for k, v in algo_params.items())
+
+ # Prepare inference environments
+ self._setups()
+
+ @torch.no_grad()
+ def conduct_prediction(self):
+ self.model.to(self.device)
+ self.model.eval()
+ total_time = 0
+ total_times = []
+
+ for img_name in self.img_names:
+ img_data = self._get_img_data(img_name)
+ img_data = img_data.to(self.device)
+
+ start = time.time()
+
+ pred_mask = self._inference(img_data)
+ pred_mask = self._post_process(pred_mask.squeeze(0).cpu().numpy())
+
+ self.write_pred_mask(
+ pred_mask, self.output_path, img_name, self.make_submission
+ )
+ self.save_prediction(
+ pred_mask, image_name=img_name)
+ end = time.time()
+
+ time_cost = end - start
+ total_times.append(time_cost)
+ total_time += time_cost
+ print(
+ f"Prediction finished: {img_name}; img size = {img_data.shape}; costing: {time_cost:.2f}s"
+ )
+
+ print(f"\n Total Time Cost: {total_time:.2f}s")
+
+ if self.make_submission:
+ fname = "%s.zip" % self.exp_name
+
+ os.makedirs("./submissions", exist_ok=True)
+ submission_path = os.path.join("./submissions", fname)
+
+ with ZipFile(submission_path, "w") as zipObj2:
+ pred_names = sorted(os.listdir(self.output_path))
+ for pred_name in pred_names:
+ pred_path = os.path.join(self.output_path, pred_name)
+ zipObj2.write(pred_path)
+
+ print("\n>>>>> Submission file is saved at: %s\n" % submission_path)
+
+ return time_cost
+
+ def write_pred_mask(self, pred_mask,
+ output_dir,
+ image_name,
+ submission=False):
+
+ # All images should contain at least 5 cells
+ if submission:
+ if not (np.max(pred_mask) > 5):
+ print("[!Caution] Only %d Cells Detected!!!\n" %
+ np.max(pred_mask))
+
+ file_name = image_name.split(".")[0]
+ file_name = file_name + "_label.tiff"
+ file_path = os.path.join(output_dir, file_name)
+
+ tif.imwrite(file_path, pred_mask, compression="zlib")
+
+ def _setups(self):
+ self.pred_transforms = get_pred_transforms()
+ os.makedirs(self.output_path, exist_ok=True)
+
+ now = datetime.now(timezone("Asia/Seoul"))
+ dt_string = now.strftime("%m%d_%H%M")
+ self.exp_name = (
+ self.exp_name + dt_string if self.exp_name is not None else dt_string
+ )
+
+ self.img_names = sorted(os.listdir(self.input_path))
+
+ def _get_img_data(self, img_name):
+ img_path = os.path.join(self.input_path, img_name)
+ img_data = self.pred_transforms(img_path)
+ img_data = img_data.unsqueeze(0)
+
+ return img_data
+
+ def _inference(self, img_data):
+ raise NotImplementedError
+
+ def _post_process(self, pred_mask):
+ raise NotImplementedError
diff --git a/annolid/segmentation/MEDIAR/core/BaseTrainer.py b/annolid/segmentation/MEDIAR/core/BaseTrainer.py
new file mode 100644
index 00000000..102f1b4e
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/BaseTrainer.py
@@ -0,0 +1,240 @@
+import torch
+import numpy as np
+from tqdm import tqdm
+from monai.inferers import sliding_window_inference
+from monai.metrics import CumulativeAverage
+from monai.transforms import (
+ Activations,
+ AsDiscrete,
+ Compose,
+ EnsureType,
+)
+
+import os, sys
+import copy
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../")))
+
+from annolid.segmentation.MEDIAR.core.utils import print_learning_device, print_with_logging
+from annolid.segmentation.MEDIAR.train_tools.measures import evaluate_f1_score_cellseg
+
+
+class BaseTrainer:
+ """Abstract base class for trainer implementations"""
+
+ def __init__(
+ self,
+ model,
+ dataloaders,
+ optimizer,
+ scheduler=None,
+ criterion=None,
+ num_epochs=100,
+ device="cuda:0",
+ no_valid=False,
+ valid_frequency=1,
+ amp=False,
+ algo_params=None,
+ ):
+ self.model = model.to(device)
+ self.dataloaders = dataloaders
+ self.optimizer = optimizer
+ self.scheduler = scheduler
+ self.criterion = criterion
+ self.num_epochs = num_epochs
+ self.no_valid = no_valid
+ self.valid_frequency = valid_frequency
+ self.device = device
+ self.amp = amp
+ self.best_weights = None
+ self.best_f1_score = 0.1
+
+ # FP-16 Scaler
+ self.scaler = torch.cuda.amp.GradScaler() if amp else None
+
+ # Assign algoritm-specific arguments
+ if algo_params:
+ self.__dict__.update((k, v) for k, v in algo_params.items())
+
+ # Cumulitive statistics
+ self.loss_metric = CumulativeAverage()
+ self.f1_metric = CumulativeAverage()
+
+ # Post-processing functions
+ self.post_pred = Compose(
+ [EnsureType(), Activations(softmax=True), AsDiscrete(threshold=0.5)]
+ )
+ self.post_gt = Compose([EnsureType(), AsDiscrete(to_onehot=None)])
+
+ def train(self):
+ """Train the model"""
+
+ # Print learning device name
+ print_learning_device(self.device)
+
+ # Learning process
+ for epoch in range(1, self.num_epochs + 1):
+ print(f"[Round {epoch}/{self.num_epochs}]")
+
+ # Train Epoch Phase
+ print(">>> Train Epoch")
+ train_results = self._epoch_phase("train")
+ print_with_logging(train_results, epoch)
+
+ if self.scheduler is not None:
+ self.scheduler.step()
+
+ if epoch % self.valid_frequency == 0:
+ if not self.no_valid:
+ # Valid Epoch Phase
+ print(">>> Valid Epoch")
+ valid_results = self._epoch_phase("valid")
+ print_with_logging(valid_results, epoch)
+
+ if "Valid_F1_Score" in valid_results.keys():
+ current_f1_score = valid_results["Valid_F1_Score"]
+ self._update_best_model(current_f1_score)
+ else:
+ print(">>> TuningSet Epoch")
+ tuning_cell_counts = self._tuningset_evaluation()
+ tuning_count_dict = {"TuningSet_Cell_Count": tuning_cell_counts}
+ print_with_logging(tuning_count_dict, epoch)
+
+ current_cell_count = tuning_cell_counts
+ self._update_best_model(current_cell_count)
+
+ print("-" * 50)
+
+ self.best_f1_score = 0
+
+ if self.best_weights is not None:
+ self.model.load_state_dict(self.best_weights)
+
+ def _epoch_phase(self, phase):
+ """Learning process for 1 Epoch (for different phases).
+
+ Args:
+ phase (str): "train", "valid", "test"
+
+ Returns:
+ dict: statistics for the phase results
+ """
+ phase_results = {}
+
+ # Set model mode
+ self.model.train() if phase == "train" else self.model.eval()
+
+ # Epoch process
+ for batch_data in tqdm(self.dataloaders[phase]):
+ images = batch_data["img"].to(self.device)
+ labels = batch_data["label"].to(self.device)
+ self.optimizer.zero_grad()
+
+ # Forward pass
+ with torch.set_grad_enabled(phase == "train"):
+ outputs = self.model(images)
+ loss = self.criterion(outputs, labels)
+ self.loss_metric.append(loss)
+
+ # Backward pass
+ if phase == "train":
+ loss.backward()
+ self.optimizer.step()
+
+ # Update metrics
+ phase_results = self._update_results(
+ phase_results, self.loss_metric, "loss", phase
+ )
+
+ return phase_results
+
+ @torch.no_grad()
+ def _tuningset_evaluation(self):
+ cell_counts_total = []
+ self.model.eval()
+
+ for batch_data in tqdm(self.dataloaders["tuning"]):
+ images = batch_data["img"].to(self.device)
+ if images.shape[-1] > 5000:
+ continue
+
+ outputs = sliding_window_inference(
+ images,
+ roi_size=512,
+ sw_batch_size=4,
+ predictor=self.model,
+ padding_mode="constant",
+ mode="gaussian",
+ )
+
+ outputs = outputs.squeeze(0)
+ outputs, _ = self._post_process(outputs, None)
+ count = len(np.unique(outputs) - 1)
+ cell_counts_total.append(count)
+
+ cell_counts_total_sum = np.sum(cell_counts_total)
+ print("Cell Counts Total: (%d)" % (cell_counts_total_sum))
+
+ return cell_counts_total_sum
+
+ def _update_results(self, phase_results, metric, metric_key, phase="train"):
+ """Aggregate and flush metrics
+
+ Args:
+ phase_results (dict): base dictionary to log metrics
+ metric (_type_): cumulated metrics
+ metric_key (_type_): name of metric
+ phase (str, optional): current phase name. Defaults to "train".
+
+ Returns:
+ dict: dictionary of metrics for the current phase
+ """
+
+ # Refine metrics name
+ metric_key = "_".join([phase, metric_key]).title()
+
+ # Aggregate metrics
+ metric_item = round(metric.aggregate().item(), 4)
+
+ # Log metrics to dictionary
+ phase_results[metric_key] = metric_item
+
+ # Flush metrics
+ metric.reset()
+
+ return phase_results
+
+ def _update_best_model(self, current_f1_score):
+ if current_f1_score > self.best_f1_score:
+ self.best_weights = copy.deepcopy(self.model.state_dict())
+ self.best_f1_score = current_f1_score
+ print(
+ "\n>>>> Update Best Model with score: {}\n".format(self.best_f1_score)
+ )
+ else:
+ pass
+
+ def _inference(self, images, phase="train"):
+ """inference methods for different phase"""
+ if phase != "train":
+ outputs = sliding_window_inference(
+ images,
+ roi_size=512,
+ sw_batch_size=4,
+ predictor=self.model,
+ padding_mode="reflect",
+ mode="gaussian",
+ overlap=0.5,
+ )
+ else:
+ outputs = self.model(images)
+
+ return outputs
+
+ def _post_process(self, outputs, labels):
+ return outputs, labels
+
+ def _get_f1_metric(self, masks_pred, masks_true):
+ f1_score = evaluate_f1_score_cellseg(masks_true, masks_pred)[-1]
+
+ return f1_score
diff --git a/annolid/segmentation/MEDIAR/core/Baseline/Predictor.py b/annolid/segmentation/MEDIAR/core/Baseline/Predictor.py
new file mode 100644
index 00000000..b4036f40
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/Baseline/Predictor.py
@@ -0,0 +1,59 @@
+import torch
+import os, sys
+from skimage import morphology, measure
+from monai.inferers import sliding_window_inference
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+from annolid.segmentation.MEDIAR.core.BasePredictor import BasePredictor
+
+__all__ = ["Predictor"]
+
+
+class Predictor(BasePredictor):
+ def __init__(
+ self,
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission=False,
+ exp_name=None,
+ algo_params=None,
+ ):
+ super(Predictor, self).__init__(
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission,
+ exp_name,
+ algo_params,
+ )
+
+ def _inference(self, img_data):
+ pred_mask = sliding_window_inference(
+ img_data,
+ 512,
+ 4,
+ self.model,
+ padding_mode="constant",
+ mode="gaussian",
+ overlap=0.6,
+ )
+
+ return pred_mask
+
+ def _post_process(self, pred_mask):
+ # Get probability map from the predicted logits
+ pred_mask = torch.from_numpy(pred_mask)
+ pred_mask = torch.softmax(pred_mask, dim=0)
+ pred_mask = pred_mask[1].cpu().numpy()
+
+ # Apply morphological post-processing
+ pred_mask = pred_mask > 0.5
+ pred_mask = morphology.remove_small_holes(pred_mask, connectivity=1)
+ pred_mask = morphology.remove_small_objects(pred_mask, 16)
+ pred_mask = measure.label(pred_mask)
+
+ return pred_mask
diff --git a/annolid/segmentation/MEDIAR/core/Baseline/Trainer.py b/annolid/segmentation/MEDIAR/core/Baseline/Trainer.py
new file mode 100644
index 00000000..1c4585e1
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/Baseline/Trainer.py
@@ -0,0 +1,115 @@
+from tqdm import tqdm
+from annolid.segmentation.MEDIAR.train_tools.measures import evaluate_f1_score_cellseg
+from annolid.segmentation.MEDIAR.core.Baseline.utils import create_interior_onehot, identify_instances_from_classmap
+from annolid.segmentation.MEDIAR.core.BaseTrainer import BaseTrainer
+import torch
+import os
+import sys
+import monai
+
+from monai.data import decollate_batch
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+
+__all__ = ["Trainer"]
+
+
+class Trainer(BaseTrainer):
+ def __init__(
+ self,
+ model,
+ dataloaders,
+ optimizer,
+ scheduler=None,
+ criterion=None,
+ num_epochs=100,
+ device="cuda:0",
+ no_valid=False,
+ valid_frequency=1,
+ amp=False,
+ algo_params=None,
+ ):
+ super(Trainer, self).__init__(
+ model,
+ dataloaders,
+ optimizer,
+ scheduler,
+ criterion,
+ num_epochs,
+ device,
+ no_valid,
+ valid_frequency,
+ amp,
+ algo_params,
+ )
+
+ # Dice loss as segmentation criterion
+ self.criterion = monai.losses.DiceCELoss(softmax=True)
+
+ def _epoch_phase(self, phase):
+ """Learning process for 1 Epoch."""
+
+ phase_results = {}
+
+ # Set model mode
+ self.model.train() if phase == "train" else self.model.eval()
+
+ # Epoch process
+ for batch_data in tqdm(self.dataloaders[phase]):
+ images = batch_data["img"].to(self.device)
+ labels = batch_data["label"].to(self.device)
+ self.optimizer.zero_grad()
+
+ # Map label masks to 3-class onehot map
+ labels_onehot = create_interior_onehot(labels)
+
+ # Forward pass
+ with torch.set_grad_enabled(phase == "train"):
+ outputs = self._inference(images, phase)
+ loss = self.criterion(outputs, labels_onehot)
+ self.loss_metric.append(loss)
+
+ if phase != "train":
+ f1_score = self._get_f1_metric(outputs, labels)
+ self.f1_metric.append(f1_score)
+
+ # Backward pass
+ if phase == "train":
+ # For the mixed precision training
+ if self.amp:
+ self.scaler.scale(loss).backward()
+ self.scaler.unscale_(self.optimizer)
+ self.scaler.step(self.optimizer)
+ self.scaler.update()
+
+ else:
+ loss.backward()
+ self.optimizer.step()
+
+ # Update metrics
+ phase_results = self._update_results(
+ phase_results, self.loss_metric, "loss", phase
+ )
+
+ if phase != "train":
+ phase_results = self._update_results(
+ phase_results, self.f1_metric, "f1_score", phase
+ )
+
+ return phase_results
+
+ def _post_process(self, outputs, labels_onehot):
+ """Conduct post-processing for outputs & labels."""
+ outputs = [self.post_pred(i) for i in decollate_batch(outputs)]
+ labels_onehot = [self.post_gt(i)
+ for i in decollate_batch(labels_onehot)]
+
+ return outputs, labels_onehot
+
+ def _get_f1_metric(self, masks_pred, masks_true):
+ masks_pred = identify_instances_from_classmap(masks_pred[0])
+ masks_true = masks_true.squeeze(0).squeeze(0).cpu().numpy()
+ f1_score = evaluate_f1_score_cellseg(masks_true, masks_pred)[-1]
+
+ return f1_score
diff --git a/annolid/segmentation/MEDIAR/core/Baseline/__init__.py b/annolid/segmentation/MEDIAR/core/Baseline/__init__.py
new file mode 100644
index 00000000..5d102375
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/Baseline/__init__.py
@@ -0,0 +1,2 @@
+from .Trainer import *
+from .Predictor import *
diff --git a/annolid/segmentation/MEDIAR/core/Baseline/utils.py b/annolid/segmentation/MEDIAR/core/Baseline/utils.py
new file mode 100644
index 00000000..4fd559de
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/Baseline/utils.py
@@ -0,0 +1,80 @@
+"""
+Adapted from the following references:
+[1] https://github.com/JunMa11/NeurIPS-CellSeg/blob/main/baseline/model_training_3class.py
+
+"""
+
+import torch
+import numpy as np
+from skimage import segmentation, morphology, measure
+import monai
+
+
+__all__ = ["create_interior_onehot", "identify_instances_from_classmap"]
+
+
+@torch.no_grad()
+def identify_instances_from_classmap(
+ class_map, cell_class=1, threshold=0.5, from_logits=True
+):
+ """Identification of cell instances from the class map"""
+
+ if from_logits:
+ class_map = torch.softmax(class_map, dim=0) # (C, H, W)
+
+ # Convert probability map to binary mask
+ pred_mask = class_map[cell_class].cpu().numpy()
+
+ # Apply morphological postprocessing
+ pred_mask = pred_mask > threshold
+ pred_mask = morphology.remove_small_holes(pred_mask, connectivity=1)
+ pred_mask = morphology.remove_small_objects(pred_mask, 16)
+ pred_mask = measure.label(pred_mask)
+
+ return pred_mask
+
+
+@torch.no_grad()
+def create_interior_onehot(inst_maps):
+ """
+ interior : (H,W), np.uint8
+ three-class map, values: 0,1,2
+ 0: background
+ 1: interior
+ 2: boundary
+ """
+ device = inst_maps.device
+
+ # Get (np.int16) array corresponding to label masks: (B, 1, H, W)
+ inst_maps = inst_maps.squeeze(1).cpu().numpy().astype(np.int16)
+
+ interior_maps = []
+
+ for inst_map in inst_maps:
+ # Create interior-edge map
+ boundary = segmentation.find_boundaries(inst_map, mode="inner")
+
+ # Refine interior-edge map
+ boundary = morphology.binary_dilation(boundary, morphology.disk(1))
+
+ # Assign label classes
+ interior_temp = np.logical_and(~boundary, inst_map > 0)
+
+ # interior_temp[boundary] = 0
+ interior_temp = morphology.remove_small_objects(interior_temp, min_size=16)
+ interior = np.zeros_like(inst_map, dtype=np.uint8)
+ interior[interior_temp] = 1
+ interior[boundary] = 2
+
+ interior_maps.append(interior)
+
+ # Aggregate interior_maps for batch
+ interior_maps = np.stack(interior_maps, axis=0).astype(np.uint8)
+
+ # Reshape as original label shape: (B, H, W)
+ interior_maps = torch.from_numpy(interior_maps).unsqueeze(1).to(device)
+
+ # Obtain one-hot map for batch
+ interior_onehot = monai.networks.one_hot(interior_maps, num_classes=3)
+
+ return interior_onehot
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/EnsemblePredictor.py b/annolid/segmentation/MEDIAR/core/MEDIAR/EnsemblePredictor.py
new file mode 100644
index 00000000..e65685c7
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/EnsemblePredictor.py
@@ -0,0 +1,105 @@
+import torch
+import os, sys, copy
+import numpy as np
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+from annolid.segmentation.MEDIAR.core.MEDIAR.Predictor import Predictor
+
+__all__ = ["EnsemblePredictor"]
+
+
+class EnsemblePredictor(Predictor):
+ def __init__(
+ self,
+ model,
+ model_aux,
+ device,
+ input_path,
+ output_path,
+ make_submission=False,
+ exp_name=None,
+ algo_params=None,
+ ):
+ super(EnsemblePredictor, self).__init__(
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission,
+ exp_name,
+ algo_params,
+ )
+ self.model_aux = model_aux
+
+ @torch.no_grad()
+ def _inference(self, img_data):
+
+ self.model_aux.to(self.device)
+ self.model_aux.eval()
+
+ img_data = img_data.to(self.device)
+ img_base = img_data
+
+ outputs_base = self._window_inference(img_base)
+ outputs_base = outputs_base.cpu().squeeze()
+
+ outputs_aux = self._window_inference(img_base, aux=True)
+ outputs_aux = outputs_aux.cpu().squeeze()
+ img_base.cpu()
+
+ if not self.use_tta:
+ pred_mask = (outputs_base + outputs_aux) / 2
+ return pred_mask
+
+ else:
+ # HorizontalFlip TTA
+ img_hflip = self.hflip_tta.apply_aug_image(img_data, apply=True)
+
+ outputs_hflip = self._window_inference(img_hflip)
+ outputs_hflip_aux = self._window_inference(img_hflip, aux=True)
+
+ outputs_hflip = self.hflip_tta.apply_deaug_mask(outputs_hflip, apply=True)
+ outputs_hflip_aux = self.hflip_tta.apply_deaug_mask(
+ outputs_hflip_aux, apply=True
+ )
+
+ outputs_hflip = outputs_hflip.cpu().squeeze()
+ outputs_hflip_aux = outputs_hflip_aux.cpu().squeeze()
+ img_hflip = img_hflip.cpu()
+
+ # VertricalFlip TTA
+ img_vflip = self.vflip_tta.apply_aug_image(img_data, apply=True)
+
+ outputs_vflip = self._window_inference(img_vflip)
+ outputs_vflip_aux = self._window_inference(img_vflip, aux=True)
+
+ outputs_vflip = self.vflip_tta.apply_deaug_mask(outputs_vflip, apply=True)
+ outputs_vflip_aux = self.vflip_tta.apply_deaug_mask(
+ outputs_vflip_aux, apply=True
+ )
+
+ outputs_vflip = outputs_vflip.cpu().squeeze()
+ outputs_vflip_aux = outputs_vflip_aux.cpu().squeeze()
+ img_vflip = img_vflip.cpu()
+
+ # Merge Results
+ pred_mask = torch.zeros_like(outputs_base)
+ pred_mask[0] = (outputs_base[0] + outputs_hflip[0] - outputs_vflip[0]) / 3
+ pred_mask[1] = (outputs_base[1] - outputs_hflip[1] + outputs_vflip[1]) / 3
+ pred_mask[2] = (outputs_base[2] + outputs_hflip[2] + outputs_vflip[2]) / 3
+
+ pred_mask_aux = torch.zeros_like(outputs_aux)
+ pred_mask_aux[0] = (
+ outputs_aux[0] + outputs_hflip_aux[0] - outputs_vflip_aux[0]
+ ) / 3
+ pred_mask_aux[1] = (
+ outputs_aux[1] - outputs_hflip_aux[1] + outputs_vflip_aux[1]
+ ) / 3
+ pred_mask_aux[2] = (
+ outputs_aux[2] + outputs_hflip_aux[2] + outputs_vflip_aux[2]
+ ) / 3
+
+ pred_mask = (pred_mask + pred_mask_aux) / 2
+
+ return pred_mask
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/Predictor.py b/annolid/segmentation/MEDIAR/core/MEDIAR/Predictor.py
new file mode 100644
index 00000000..da3f12eb
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/Predictor.py
@@ -0,0 +1,234 @@
+import torch
+import numpy as np
+import os, sys
+from monai.inferers import sliding_window_inference
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+from annolid.segmentation.MEDIAR.core.BasePredictor import BasePredictor
+from annolid.segmentation.MEDIAR.core.MEDIAR.utils import compute_masks
+
+__all__ = ["Predictor"]
+
+
+class Predictor(BasePredictor):
+ def __init__(
+ self,
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission=False,
+ exp_name=None,
+ algo_params=None,
+ ):
+ super(Predictor, self).__init__(
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission,
+ exp_name,
+ algo_params,
+ )
+ self.hflip_tta = HorizontalFlip()
+ self.vflip_tta = VerticalFlip()
+
+ @torch.no_grad()
+ def _inference(self, img_data):
+ """Conduct model prediction"""
+
+ img_data = img_data.to(self.device)
+ img_base = img_data
+ outputs_base = self._window_inference(img_base)
+ outputs_base = outputs_base.cpu().squeeze()
+ img_base.cpu()
+
+ if not self.use_tta:
+ pred_mask = outputs_base
+ return pred_mask
+
+ else:
+ # HorizontalFlip TTA
+ img_hflip = self.hflip_tta.apply_aug_image(img_data, apply=True)
+ outputs_hflip = self._window_inference(img_hflip)
+ outputs_hflip = self.hflip_tta.apply_deaug_mask(outputs_hflip, apply=True)
+ outputs_hflip = outputs_hflip.cpu().squeeze()
+ img_hflip = img_hflip.cpu()
+
+ # VertricalFlip TTA
+ img_vflip = self.vflip_tta.apply_aug_image(img_data, apply=True)
+ outputs_vflip = self._window_inference(img_vflip)
+ outputs_vflip = self.vflip_tta.apply_deaug_mask(outputs_vflip, apply=True)
+ outputs_vflip = outputs_vflip.cpu().squeeze()
+ img_vflip = img_vflip.cpu()
+
+ # Merge Results
+ pred_mask = torch.zeros_like(outputs_base)
+ pred_mask[0] = (outputs_base[0] + outputs_hflip[0] - outputs_vflip[0]) / 3
+ pred_mask[1] = (outputs_base[1] - outputs_hflip[1] + outputs_vflip[1]) / 3
+ pred_mask[2] = (outputs_base[2] + outputs_hflip[2] + outputs_vflip[2]) / 3
+
+ return pred_mask
+
+ def _window_inference(self, img_data, aux=False):
+ """Inference on RoI-sized window"""
+ outputs = sliding_window_inference(
+ img_data,
+ roi_size=512,
+ sw_batch_size=4,
+ predictor=self.model if not aux else self.model_aux,
+ padding_mode="constant",
+ mode="gaussian",
+ overlap=0.6,
+ )
+
+ return outputs
+
+ def _post_process(self, pred_mask):
+ """Generate cell instance masks."""
+ dP, cellprob = pred_mask[:2], self._sigmoid(pred_mask[-1])
+ H, W = pred_mask.shape[-2], pred_mask.shape[-1]
+
+ if np.prod(H * W) < (5000 * 5000):
+ pred_mask = compute_masks(
+ dP,
+ cellprob,
+ use_gpu=True,
+ flow_threshold=0.4,
+ device=self.device,
+ cellprob_threshold=0.5,
+ )[0]
+
+ else:
+ print("\n[Whole Slide] Grid Prediction starting...")
+ roi_size = 2000
+
+ # Get patch grid by roi_size
+ if H % roi_size != 0:
+ n_H = H // roi_size + 1
+ new_H = roi_size * n_H
+ else:
+ n_H = H // roi_size
+ new_H = H
+
+ if W % roi_size != 0:
+ n_W = W // roi_size + 1
+ new_W = roi_size * n_W
+ else:
+ n_W = W // roi_size
+ new_W = W
+
+ # Allocate values on the grid
+ pred_pad = np.zeros((new_H, new_W), dtype=np.uint32)
+ dP_pad = np.zeros((2, new_H, new_W), dtype=np.float32)
+ cellprob_pad = np.zeros((new_H, new_W), dtype=np.float32)
+
+ dP_pad[:, :H, :W], cellprob_pad[:H, :W] = dP, cellprob
+
+ for i in range(n_H):
+ for j in range(n_W):
+ print("Pred on Grid (%d, %d) processing..." % (i, j))
+ dP_roi = dP_pad[
+ :,
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ]
+ cellprob_roi = cellprob_pad[
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ]
+
+ pred_mask = compute_masks(
+ dP_roi,
+ cellprob_roi,
+ use_gpu=True,
+ flow_threshold=0.4,
+ device=self.device,
+ cellprob_threshold=0.5,
+ )[0]
+
+ pred_pad[
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ] = pred_mask
+
+ pred_mask = pred_pad[:H, :W]
+
+ return pred_mask
+
+ def _sigmoid(self, z):
+ return 1 / (1 + np.exp(-z))
+
+
+"""
+Adapted from the following references:
+[1] https://github.com/qubvel/ttach/blob/master/ttach/transforms.py
+
+"""
+
+
+def hflip(x):
+ """flip batch of images horizontally"""
+ return x.flip(3)
+
+
+def vflip(x):
+ """flip batch of images vertically"""
+ return x.flip(2)
+
+
+class DualTransform:
+ identity_param = None
+
+ def __init__(
+ self, name: str, params,
+ ):
+ self.params = params
+ self.pname = name
+
+ def apply_aug_image(self, image, *args, **params):
+ raise NotImplementedError
+
+ def apply_deaug_mask(self, mask, *args, **params):
+ raise NotImplementedError
+
+
+class HorizontalFlip(DualTransform):
+ """Flip images horizontally (left -> right)"""
+
+ identity_param = False
+
+ def __init__(self):
+ super().__init__("apply", [False, True])
+
+ def apply_aug_image(self, image, apply=False, **kwargs):
+ if apply:
+ image = hflip(image)
+ return image
+
+ def apply_deaug_mask(self, mask, apply=False, **kwargs):
+ if apply:
+ mask = hflip(mask)
+ return mask
+
+
+class VerticalFlip(DualTransform):
+ """Flip images vertically (up -> down)"""
+
+ identity_param = False
+
+ def __init__(self):
+ super().__init__("apply", [False, True])
+
+ def apply_aug_image(self, image, apply=False, **kwargs):
+ if apply:
+ image = vflip(image)
+
+ return image
+
+ def apply_deaug_mask(self, mask, apply=False, **kwargs):
+ if apply:
+ mask = vflip(mask)
+
+ return mask
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/Trainer.py b/annolid/segmentation/MEDIAR/core/MEDIAR/Trainer.py
new file mode 100644
index 00000000..4661f339
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/Trainer.py
@@ -0,0 +1,172 @@
+import torch
+import torch.nn as nn
+import numpy as np
+import os, sys
+from tqdm import tqdm
+from monai.inferers import sliding_window_inference
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+from annolid.segmentation.MEDIAR.core.BaseTrainer import BaseTrainer
+from annolid.segmentation.MEDIAR.core.MEDIAR.utils import *
+
+__all__ = ["Trainer"]
+
+
+class Trainer(BaseTrainer):
+ def __init__(
+ self,
+ model,
+ dataloaders,
+ optimizer,
+ scheduler=None,
+ criterion=None,
+ num_epochs=100,
+ device="cuda:0",
+ no_valid=False,
+ valid_frequency=1,
+ amp=False,
+ algo_params=None,
+ ):
+ super(Trainer, self).__init__(
+ model,
+ dataloaders,
+ optimizer,
+ scheduler,
+ criterion,
+ num_epochs,
+ device,
+ no_valid,
+ valid_frequency,
+ amp,
+ algo_params,
+ )
+
+ self.mse_loss = nn.MSELoss(reduction="mean")
+ self.bce_loss = nn.BCEWithLogitsLoss(reduction="mean")
+
+ def mediar_criterion(self, outputs, labels_onehot_flows):
+ """loss function between true labels and prediction outputs"""
+
+ # Cell Recognition Loss
+ cellprob_loss = self.bce_loss(
+ outputs[:, -1],
+ torch.from_numpy(labels_onehot_flows[:, 1] > 0.5).to(self.device).float(),
+ )
+
+ # Cell Distinction Loss
+ gradient_flows = torch.from_numpy(labels_onehot_flows[:, 2:]).to(self.device)
+ gradflow_loss = 0.5 * self.mse_loss(outputs[:, :2], 5.0 * gradient_flows)
+
+ loss = cellprob_loss + gradflow_loss
+
+ return loss
+
+ def _epoch_phase(self, phase):
+ phase_results = {}
+
+ # Set model mode
+ self.model.train() if phase == "train" else self.model.eval()
+
+ # Epoch process
+ for batch_data in tqdm(self.dataloaders[phase]):
+ images, labels = batch_data["img"], batch_data["label"]
+
+ if self.with_public:
+ # Load batches sequentially from the unlabeled dataloader
+ try:
+ batch_data = next(self.public_iterator)
+ images_pub, labels_pub = batch_data["img"], batch_data["label"]
+
+ except:
+ # Assign memory loader if the cycle ends
+ self.public_iterator = iter(self.public_loader)
+ batch_data = next(self.public_iterator)
+ images_pub, labels_pub = batch_data["img"], batch_data["label"]
+
+ # Concat memory data to the batch
+ images = torch.cat([images, images_pub], dim=0)
+ labels = torch.cat([labels, labels_pub], dim=0)
+
+ images = images.to(self.device)
+ labels = labels.to(self.device)
+
+ self.optimizer.zero_grad()
+
+ # Forward pass
+ with torch.cuda.amp.autocast(enabled=self.amp):
+ with torch.set_grad_enabled(phase == "train"):
+ # Output shape is B x [grad y, grad x, cellprob] x H x W
+ outputs = self._inference(images, phase)
+
+ # Map label masks to graidnet and onehot
+ labels_onehot_flows = labels_to_flows(
+ labels, use_gpu=True, device=self.device
+ )
+ # Calculate loss
+ loss = self.mediar_criterion(outputs, labels_onehot_flows)
+ self.loss_metric.append(loss)
+
+ # Calculate valid statistics
+ if phase != "train":
+ outputs, labels = self._post_process(outputs, labels)
+ f1_score = self._get_f1_metric(outputs, labels)
+ self.f1_metric.append(f1_score)
+
+ # Backward pass
+ if phase == "train":
+ # For the mixed precision training
+ if self.amp:
+ self.scaler.scale(loss).backward()
+ self.scaler.unscale_(self.optimizer)
+ self.scaler.step(self.optimizer)
+ self.scaler.update()
+
+ else:
+ loss.backward()
+ self.optimizer.step()
+
+ # Update metrics
+ phase_results = self._update_results(
+ phase_results, self.loss_metric, "dice_loss", phase
+ )
+ if phase != "train":
+ phase_results = self._update_results(
+ phase_results, self.f1_metric, "f1_score", phase
+ )
+
+ return phase_results
+
+ def _inference(self, images, phase="train"):
+ """inference methods for different phase"""
+
+ if phase != "train":
+ outputs = sliding_window_inference(
+ images,
+ roi_size=512,
+ sw_batch_size=4,
+ predictor=self.model,
+ padding_mode="constant",
+ mode="gaussian",
+ overlap=0.5,
+ )
+ else:
+ outputs = self.model(images)
+
+ return outputs
+
+ def _post_process(self, outputs, labels=None):
+ """Predict cell instances using the gradient tracking"""
+ outputs = outputs.squeeze(0).cpu().numpy()
+ gradflows, cellprob = outputs[:2], self._sigmoid(outputs[-1])
+ outputs = compute_masks(gradflows, cellprob, use_gpu=True, device=self.device)
+ outputs = outputs[0] # (1, C, H, W) -> (C, H, W)
+
+ if labels is not None:
+ labels = labels.squeeze(0).squeeze(0).cpu().numpy()
+
+ return outputs, labels
+
+ def _sigmoid(self, z):
+ """Sigmoid function for numpy arrays"""
+ return 1 / (1 + np.exp(-z))
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/__init__.py b/annolid/segmentation/MEDIAR/core/MEDIAR/__init__.py
new file mode 100644
index 00000000..80cc69b9
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/__init__.py
@@ -0,0 +1,3 @@
+from .Trainer import *
+from .Predictor import *
+from .EnsemblePredictor import *
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/utils.py b/annolid/segmentation/MEDIAR/core/MEDIAR/utils.py
new file mode 100644
index 00000000..555ea4e1
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/utils.py
@@ -0,0 +1,429 @@
+"""
+Copyright © 2022 Howard Hughes Medical Institute,
+Authored by Carsen Stringer and Marius Pachitariu.
+
+Redistribution and use in source and binary forms, with or without
+modification, are permitted provided that the following conditions are met:
+
+1. Redistributions of source code must retain the above copyright notice,
+ this list of conditions and the following disclaimer.
+
+2. Redistributions in binary form must reproduce the above copyright notice,
+ this list of conditions and the following disclaimer in the documentation
+ and/or other materials provided with the distribution.
+
+3. Neither the name of HHMI nor the names of its contributors may be used to
+ endorse or promote products derived from this software without specific
+ prior written permission.
+
+THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
+AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
+IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE
+ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT OWNER OR CONTRIBUTORS BE
+LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR
+CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF
+SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS
+INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN
+CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE)
+ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE
+POSSIBILITY OF SUCH DAMAGE.
+
+--------------------------------------------------------------------------
+MEDIAR Prediction uses CellPose's Gradient Flow Tracking.
+
+This code is adapted from the following codes:
+[1] https://github.com/MouseLand/cellpose/blob/main/cellpose/utils.py
+[2] https://github.com/MouseLand/cellpose/blob/main/cellpose/dynamics.py
+[3] https://github.com/MouseLand/cellpose/blob/main/cellpose/metrics.py
+"""
+
+import torch
+from torch.nn.functional import grid_sample
+import numpy as np
+import fastremap
+
+from skimage import morphology
+from scipy.ndimage import mean, find_objects
+from scipy.ndimage.filters import maximum_filter1d
+
+torch_GPU = torch.device("cuda")
+torch_CPU = torch.device("cpu")
+
+
+def labels_to_flows(labels, use_gpu=False, device=None, redo_flows=False):
+ """
+ Convert labels (list of masks or flows) to flows for training model
+ """
+
+ # Labels b x 1 x h x w
+ labels = labels.cpu().numpy().astype(np.int16)
+ nimg = len(labels)
+
+ if labels[0].ndim < 3:
+ labels = [labels[n][np.newaxis, :, :] for n in range(nimg)]
+
+ # Flows need to be recomputed
+ if labels[0].shape[0] == 1 or labels[0].ndim < 3 or redo_flows:
+ # compute flows; labels are fixed here to be unique, so they need to be passed back
+ # make sure labels are unique!
+ labels = [fastremap.renumber(label, in_place=True)[0] for label in labels]
+ veci = [
+ masks_to_flows(labels[n][0], use_gpu=use_gpu, device=device)
+ for n in range(nimg)
+ ]
+
+ # concatenate labels, distance transform, vector flows, heat (boundary and mask are computed in augmentations)
+ flows = [
+ np.concatenate((labels[n], labels[n] > 0.5, veci[n]), axis=0).astype(
+ np.float32
+ )
+ for n in range(nimg)
+ ]
+
+ return np.array(flows)
+
+
+def compute_masks(
+ dP,
+ cellprob,
+ p=None,
+ niter=200,
+ cellprob_threshold=0.4,
+ flow_threshold=0.4,
+ interp=True,
+ resize=None,
+ use_gpu=False,
+ device=None,
+):
+ """compute masks using dynamics from dP, cellprob, and boundary"""
+
+ cp_mask = cellprob > cellprob_threshold
+ cp_mask = morphology.remove_small_holes(cp_mask, area_threshold=16)
+ cp_mask = morphology.remove_small_objects(cp_mask, min_size=16)
+
+ if np.any(cp_mask): # mask at this point is a cell cluster binary map, not labels
+ # follow flows
+ if p is None:
+ p, inds = follow_flows(
+ dP * cp_mask / 5.0,
+ niter=niter,
+ interp=interp,
+ use_gpu=use_gpu,
+ device=device,
+ )
+ if inds is None:
+ shape = resize if resize is not None else cellprob.shape
+ mask = np.zeros(shape, np.uint16)
+ p = np.zeros((len(shape), *shape), np.uint16)
+ return mask, p
+
+ # calculate masks
+ mask = get_masks(p, iscell=cp_mask)
+
+ # flow thresholding factored out of get_masks
+ shape0 = p.shape[1:]
+ if mask.max() > 0 and flow_threshold is not None and flow_threshold > 0:
+ # make sure labels are unique at output of get_masks
+ mask = remove_bad_flow_masks(
+ mask, dP, threshold=flow_threshold, use_gpu=use_gpu, device=device
+ )
+ else: # nothing to compute, just make it compatible
+ shape = resize if resize is not None else cellprob.shape
+ mask = np.zeros(shape, np.uint16)
+ p = np.zeros((len(shape), *shape), np.uint16)
+
+ return mask, p
+
+
+def _extend_centers_gpu(
+ neighbors, centers, isneighbor, Ly, Lx, n_iter=200, device=torch.device("cuda")
+):
+ if device is not None:
+ device = device
+ nimg = neighbors.shape[0] // 9
+ pt = torch.from_numpy(neighbors).to(device)
+
+ T = torch.zeros((nimg, Ly, Lx), dtype=torch.double, device=device)
+ meds = torch.from_numpy(centers.astype(int)).to(device).long()
+ isneigh = torch.from_numpy(isneighbor).to(device)
+ for i in range(n_iter):
+ T[:, meds[:, 0], meds[:, 1]] += 1
+ Tneigh = T[:, pt[:, :, 0], pt[:, :, 1]]
+ Tneigh *= isneigh
+ T[:, pt[0, :, 0], pt[0, :, 1]] = Tneigh.mean(axis=1)
+ del meds, isneigh, Tneigh
+ T = torch.log(1.0 + T)
+ # gradient positions
+ grads = T[:, pt[[2, 1, 4, 3], :, 0], pt[[2, 1, 4, 3], :, 1]]
+ del pt
+ dy = grads[:, 0] - grads[:, 1]
+ dx = grads[:, 2] - grads[:, 3]
+ del grads
+ mu_torch = np.stack((dy.cpu().squeeze(), dx.cpu().squeeze()), axis=-2)
+ return mu_torch
+
+
+def diameters(masks):
+ _, counts = np.unique(np.int32(masks), return_counts=True)
+ counts = counts[1:]
+ md = np.median(counts ** 0.5)
+ if np.isnan(md):
+ md = 0
+ md /= (np.pi ** 0.5) / 2
+ return md, counts ** 0.5
+
+
+def masks_to_flows_gpu(masks, device=None):
+ if device is None:
+ device = torch.device("cuda")
+
+ Ly0, Lx0 = masks.shape
+ Ly, Lx = Ly0 + 2, Lx0 + 2
+
+ masks_padded = np.zeros((Ly, Lx), np.int64)
+ masks_padded[1:-1, 1:-1] = masks
+
+ # get mask pixel neighbors
+ y, x = np.nonzero(masks_padded)
+ neighborsY = np.stack((y, y - 1, y + 1, y, y, y - 1, y - 1, y + 1, y + 1), axis=0)
+ neighborsX = np.stack((x, x, x, x - 1, x + 1, x - 1, x + 1, x - 1, x + 1), axis=0)
+ neighbors = np.stack((neighborsY, neighborsX), axis=-1)
+
+ # get mask centers
+ slices = find_objects(masks)
+
+ centers = np.zeros((masks.max(), 2), "int")
+ for i, si in enumerate(slices):
+ if si is not None:
+ sr, sc = si
+
+ ly, lx = sr.stop - sr.start + 1, sc.stop - sc.start + 1
+ yi, xi = np.nonzero(masks[sr, sc] == (i + 1))
+ yi = yi.astype(np.int32) + 1 # add padding
+ xi = xi.astype(np.int32) + 1 # add padding
+ ymed = np.median(yi)
+ xmed = np.median(xi)
+ imin = np.argmin((xi - xmed) ** 2 + (yi - ymed) ** 2)
+ xmed = xi[imin]
+ ymed = yi[imin]
+ centers[i, 0] = ymed + sr.start
+ centers[i, 1] = xmed + sc.start
+
+ # get neighbor validator (not all neighbors are in same mask)
+ neighbor_masks = masks_padded[neighbors[:, :, 0], neighbors[:, :, 1]]
+ isneighbor = neighbor_masks == neighbor_masks[0]
+ ext = np.array(
+ [[sr.stop - sr.start + 1, sc.stop - sc.start + 1] for sr, sc in slices]
+ )
+ n_iter = 2 * (ext.sum(axis=1)).max()
+ # run diffusion
+ mu = _extend_centers_gpu(
+ neighbors, centers, isneighbor, Ly, Lx, n_iter=n_iter, device=device
+ )
+
+ # normalize
+ mu /= 1e-20 + (mu ** 2).sum(axis=0) ** 0.5
+
+ # put into original image
+ mu0 = np.zeros((2, Ly0, Lx0))
+ mu0[:, y - 1, x - 1] = mu
+ mu_c = np.zeros_like(mu0)
+ return mu0, mu_c
+
+
+def masks_to_flows(masks, use_gpu=False, device=None):
+ if masks.max() == 0 or (masks != 0).sum() == 1:
+ # dynamics_logger.warning('empty masks!')
+ return np.zeros((2, *masks.shape), "float32")
+
+ if use_gpu:
+ if use_gpu and device is None:
+ device = torch_GPU
+ elif device is None:
+ device = torch_CPU
+ masks_to_flows_device = masks_to_flows_gpu
+
+ if masks.ndim == 3:
+ Lz, Ly, Lx = masks.shape
+ mu = np.zeros((3, Lz, Ly, Lx), np.float32)
+ for z in range(Lz):
+ mu0 = masks_to_flows_device(masks[z], device=device)[0]
+ mu[[1, 2], z] += mu0
+ for y in range(Ly):
+ mu0 = masks_to_flows_device(masks[:, y], device=device)[0]
+ mu[[0, 2], :, y] += mu0
+ for x in range(Lx):
+ mu0 = masks_to_flows_device(masks[:, :, x], device=device)[0]
+ mu[[0, 1], :, :, x] += mu0
+ return mu
+ elif masks.ndim == 2:
+ mu, mu_c = masks_to_flows_device(masks, device=device)
+ return mu
+
+ else:
+ raise ValueError("masks_to_flows only takes 2D or 3D arrays")
+
+
+def steps2D_interp(p, dP, niter, use_gpu=False, device=None):
+ shape = dP.shape[1:]
+ if use_gpu:
+ if device is None:
+ device = torch_GPU
+ shape = (
+ np.array(shape)[[1, 0]].astype("float") - 1
+ ) # Y and X dimensions (dP is 2.Ly.Lx), flipped X-1, Y-1
+ pt = (
+ torch.from_numpy(p[[1, 0]].T).float().to(device).unsqueeze(0).unsqueeze(0)
+ ) # p is n_points by 2, so pt is [1 1 2 n_points]
+ im = (
+ torch.from_numpy(dP[[1, 0]]).float().to(device).unsqueeze(0)
+ ) # covert flow numpy array to tensor on GPU, add dimension
+ # normalize pt between 0 and 1, normalize the flow
+ for k in range(2):
+ im[:, k, :, :] *= 2.0 / shape[k]
+ pt[:, :, :, k] /= shape[k]
+
+ # normalize to between -1 and 1
+ pt = pt * 2 - 1
+
+ # here is where the stepping happens
+ for t in range(niter):
+ # align_corners default is False, just added to suppress warning
+ dPt = grid_sample(im, pt, align_corners=False)
+
+ for k in range(2): # clamp the final pixel locations
+ pt[:, :, :, k] = torch.clamp(
+ pt[:, :, :, k] + dPt[:, k, :, :], -1.0, 1.0
+ )
+
+ # undo the normalization from before, reverse order of operations
+ pt = (pt + 1) * 0.5
+ for k in range(2):
+ pt[:, :, :, k] *= shape[k]
+
+ p = pt[:, :, :, [1, 0]].cpu().numpy().squeeze().T
+ return p
+
+ else:
+ assert print("ho")
+
+
+def follow_flows(dP, mask=None, niter=200, interp=True, use_gpu=True, device=None):
+ shape = np.array(dP.shape[1:]).astype(np.int32)
+ niter = np.uint32(niter)
+
+ p = np.meshgrid(np.arange(shape[0]), np.arange(shape[1]), indexing="ij")
+ p = np.array(p).astype(np.float32)
+
+ inds = np.array(np.nonzero(np.abs(dP[0]) > 1e-3)).astype(np.int32).T
+
+ if inds.ndim < 2 or inds.shape[0] < 5:
+ return p, None
+
+ if not interp:
+ assert print("woo")
+
+ else:
+ p_interp = steps2D_interp(
+ p[:, inds[:, 0], inds[:, 1]], dP, niter, use_gpu=use_gpu, device=device
+ )
+ p[:, inds[:, 0], inds[:, 1]] = p_interp
+
+ return p, inds
+
+
+def flow_error(maski, dP_net, use_gpu=False, device=None):
+ if dP_net.shape[1:] != maski.shape:
+ print("ERROR: net flow is not same size as predicted masks")
+ return
+
+ # flows predicted from estimated masks
+ dP_masks = masks_to_flows(maski, use_gpu=use_gpu, device=device)
+ # difference between predicted flows vs mask flows
+ flow_errors = np.zeros(maski.max())
+ for i in range(dP_masks.shape[0]):
+ flow_errors += mean(
+ (dP_masks[i] - dP_net[i] / 5.0) ** 2,
+ maski,
+ index=np.arange(1, maski.max() + 1),
+ )
+
+ return flow_errors, dP_masks
+
+
+def remove_bad_flow_masks(masks, flows, threshold=0.4, use_gpu=False, device=None):
+ merrors, _ = flow_error(masks, flows, use_gpu, device)
+ badi = 1 + (merrors > threshold).nonzero()[0]
+ masks[np.isin(masks, badi)] = 0
+ return masks
+
+
+def get_masks(p, iscell=None, rpad=20):
+ pflows = []
+ edges = []
+ shape0 = p.shape[1:]
+ dims = len(p)
+
+ for i in range(dims):
+ pflows.append(p[i].flatten().astype("int32"))
+ edges.append(np.arange(-0.5 - rpad, shape0[i] + 0.5 + rpad, 1))
+
+ h, _ = np.histogramdd(tuple(pflows), bins=edges)
+ hmax = h.copy()
+ for i in range(dims):
+ hmax = maximum_filter1d(hmax, 5, axis=i)
+
+ seeds = np.nonzero(np.logical_and(h - hmax > -1e-6, h > 10))
+ Nmax = h[seeds]
+ isort = np.argsort(Nmax)[::-1]
+ for s in seeds:
+ s = s[isort]
+
+ pix = list(np.array(seeds).T)
+
+ shape = h.shape
+ if dims == 3:
+ expand = np.nonzero(np.ones((3, 3, 3)))
+ else:
+ expand = np.nonzero(np.ones((3, 3)))
+ for e in expand:
+ e = np.expand_dims(e, 1)
+
+ for iter in range(5):
+ for k in range(len(pix)):
+ if iter == 0:
+ pix[k] = list(pix[k])
+ newpix = []
+ iin = []
+ for i, e in enumerate(expand):
+ epix = e[:, np.newaxis] + np.expand_dims(pix[k][i], 0) - 1
+ epix = epix.flatten()
+ iin.append(np.logical_and(epix >= 0, epix < shape[i]))
+ newpix.append(epix)
+ iin = np.all(tuple(iin), axis=0)
+ for p in newpix:
+ p = p[iin]
+ newpix = tuple(newpix)
+ igood = h[newpix] > 2
+ for i in range(dims):
+ pix[k][i] = newpix[i][igood]
+ if iter == 4:
+ pix[k] = tuple(pix[k])
+
+ M = np.zeros(h.shape, np.uint32)
+ for k in range(len(pix)):
+ M[pix[k]] = 1 + k
+
+ for i in range(dims):
+ pflows[i] = pflows[i] + rpad
+ M0 = M[tuple(pflows)]
+
+ # remove big masks
+ uniq, counts = fastremap.unique(M0, return_counts=True)
+ big = np.prod(shape0) * 0.9
+ bigc = uniq[counts > big]
+ if len(bigc) > 0 and (len(bigc) > 1 or bigc[0] != 0):
+ M0 = fastremap.mask(M0, bigc)
+ fastremap.renumber(M0, in_place=True) # convenient to guarantee non-skipped labels
+ M0 = np.reshape(M0, shape0)
+ return M0
diff --git a/annolid/segmentation/MEDIAR/core/__init__.py b/annolid/segmentation/MEDIAR/core/__init__.py
new file mode 100644
index 00000000..ff516dce
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/__init__.py
@@ -0,0 +1,2 @@
+from .Baseline import *
+from .MEDIAR import *
diff --git a/annolid/segmentation/MEDIAR/core/utils.py b/annolid/segmentation/MEDIAR/core/utils.py
new file mode 100644
index 00000000..5dc93fbc
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/utils.py
@@ -0,0 +1,40 @@
+import torch
+#import wandb
+import pprint
+
+__all__ = ["print_learning_device", "print_with_logging"]
+
+
+def print_learning_device(device):
+ """Get and print the learning device information."""
+ if device == "cpu":
+ device_name = device
+
+ else:
+ if isinstance(device, str):
+ device_idx = int(device[-1])
+ elif isinstance(device, torch._device):
+ device_idx = device.index
+
+ device_name = torch.cuda.get_device_name(device_idx)
+
+ print("")
+ print("=" * 50)
+ print("Train start on device: {}".format(device_name))
+ print("=" * 50)
+
+
+def print_with_logging(results, step):
+ """Print and log on the W&B server.
+
+ Args:
+ results (dict): results dictionary
+ step (int): epoch index
+ """
+ # Print the results dictionary
+ pp = pprint.PrettyPrinter(compact=True)
+ pp.pprint(results)
+ print()
+
+ # Log on the w&b server
+ #wandb.log(results, step=step)
diff --git a/annolid/segmentation/MEDIAR/download_weights.py b/annolid/segmentation/MEDIAR/download_weights.py
new file mode 100644
index 00000000..649d99a1
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/download_weights.py
@@ -0,0 +1,26 @@
+import os
+import gdown
+
+
+def download_weights(output_dir="./weights"):
+ os.makedirs(output_dir, exist_ok=True)
+
+ weights_urls = {
+ "from_phase1": "https://drive.google.com/uc?id=168MtudjTMLoq9YGTyoD2Rjl_d3Gy6c_L",
+ "from_phase2": "https://drive.google.com/uc?id=1JJ2-QKTCk-G7sp5ddkqcifMxgnyOrXjx"
+ }
+
+ for name, url in weights_urls.items():
+ output_path = os.path.join(output_dir, f"{name}.pth")
+ gdown.download(url, output_path, quiet=False)
+
+ return {
+ "model_path1": os.path.join(output_dir, "from_phase1.pth"),
+ "model_path2": os.path.join(output_dir, "from_phase2.pth")
+ }
+
+
+# Example usage:
+weights_paths = download_weights()
+# model_path1 = weights_paths["model_path1"]
+# model_path2 = weights_paths["model_path2"]
diff --git a/annolid/segmentation/MEDIAR/evaluate.py b/annolid/segmentation/MEDIAR/evaluate.py
new file mode 100644
index 00000000..b2a15930
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/evaluate.py
@@ -0,0 +1,71 @@
+import numpy as np
+import pandas as pd
+import tifffile as tif
+import argparse
+import os
+from collections import OrderedDict
+from tqdm import tqdm
+
+from annolid.segmentation.MEDIAR.train_tools.measures import evaluate_f1_score_cellseg
+
+
+def main():
+ ### Directory path arguments ###
+ parser = argparse.ArgumentParser("Compute F1 score for cell segmentation results")
+ parser.add_argument(
+ "--gt_path",
+ type=str,
+ help="path to ground truth; file names end with _label.tiff",
+ required=True,
+ )
+ parser.add_argument(
+ "--pred_path", type=str, help="path to segmentation results", required=True
+ )
+ parser.add_argument("--save_path", default=None, help="path where to save metrics")
+
+ args = parser.parse_args()
+
+ # Get files from the paths
+ gt_path, pred_path = args.gt_path, args.pred_path
+ names = sorted(os.listdir(pred_path))
+
+ names_total = []
+ precisions_total, recalls_total, f1_scores_total = [], [], []
+
+ for name in tqdm(names):
+ assert name.endswith(
+ "_label.tiff"
+ ), "The suffix of label name should be _label.tiff"
+
+ # Load the images
+ gt = tif.imread(os.path.join(gt_path, name))
+ pred = tif.imread(os.path.join(pred_path, name))
+
+ # Evaluate metrics
+ precision, recall, f1_score = evaluate_f1_score_cellseg(gt, pred, threshold=0.5)
+
+ names_total.append(name)
+ precisions_total.append(np.round(precision, 4))
+ recalls_total.append(np.round(recall, 4))
+ f1_scores_total.append(np.round(f1_score, 4))
+
+ # Refine data as dataframe
+ cellseg_metric = OrderedDict()
+ cellseg_metric["Names"] = names_total
+ cellseg_metric["Precision"] = precisions_total
+ cellseg_metric["Recall"] = recalls_total
+ cellseg_metric["F1_Score"] = f1_scores_total
+
+ cellseg_metric = pd.DataFrame(cellseg_metric)
+ print("mean F1 Score:", np.mean(cellseg_metric["F1_Score"]))
+
+ # Save results
+ if args.save_path is not None:
+ os.makedirs(args.save_path, exist_ok=True)
+ cellseg_metric.to_csv(
+ os.path.join(args.save_path, "seg_metric.csv"), index=False
+ )
+
+
+if __name__ == "__main__":
+ main()
diff --git a/annolid/segmentation/MEDIAR/generate_mapping.py b/annolid/segmentation/MEDIAR/generate_mapping.py
new file mode 100644
index 00000000..91c166d2
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/generate_mapping.py
@@ -0,0 +1,121 @@
+import os, glob
+import json
+import argparse
+
+
+def public_paths_labeled(root):
+ """Map paths for public datasets as dictionary list"""
+
+ images_raw = sorted(glob.glob(os.path.join(root, "Public/images/*")))
+ labels_raw = sorted(glob.glob(os.path.join(root, "Public/labels/*")))
+
+ data_dicts = []
+
+ for image_path, label_path in zip(images_raw, labels_raw):
+ name1 = image_path.split("/")[-1].split(".")[0]
+ name2 = label_path.split("/")[-1].split("_label")[0]
+ assert name1 == name2
+
+ data_item = {
+ "img": image_path.split("MEDIAR/")[-1],
+ "label": label_path.split("MEDIAR/")[-1],
+ }
+
+ data_dicts.append(data_item)
+
+ map_dict = {"public": data_dicts}
+
+ return map_dict
+
+
+def official_paths_labeled(root):
+ """Map paths for official labeled datasets as dictionary list"""
+
+ image_path = os.path.join(root, "Official/Training/images/*")
+ label_path = os.path.join(root, "Official/Training/labels/*")
+
+ images_raw = sorted(glob.glob(image_path))
+ labels_raw = sorted(glob.glob(label_path))
+ data_dicts = []
+
+ for image_path, label_path in zip(images_raw, labels_raw):
+ name1 = image_path.split("/")[-1].split(".")[0]
+ name2 = label_path.split("/")[-1].split("_label")[0]
+ assert name1 == name2
+
+ data_item = {
+ "img": image_path.split("MEDIAR/")[-1],
+ "label": label_path.split("MEDIAR/")[-1],
+ }
+
+ data_dicts.append(data_item)
+
+ map_dict = {"official": data_dicts}
+
+ return map_dict
+
+
+def official_paths_tuning(root):
+ """Map paths for official tuning datasets as dictionary list"""
+
+ image_path = os.path.join(root, "Official/Tuning/images/*")
+ images_raw = sorted(glob.glob(image_path))
+
+ data_dicts = []
+
+ for image_path in images_raw:
+ data_item = {"img": image_path.split("MEDIAR/")[-1]}
+ data_dicts.append(data_item)
+
+ map_dict = {"official": data_dicts}
+
+ return map_dict
+
+
+def add_mapping_to_json(json_file, map_dict):
+ """Save mapped dictionary as a json file"""
+
+ if not os.path.exists(json_file):
+ with open(json_file, "w") as file:
+ json.dump({}, file)
+
+ with open(json_file, "r") as file:
+ data = json.load(file)
+
+ for map_key, map_item in map_dict.items():
+ if map_key not in data.keys():
+ data[map_key] = map_item
+ else:
+ print('>>> "{}" already exists in path map keys...'.format(map_key))
+
+ with open(json_file, "w") as file:
+ json.dump(data, file)
+
+
+if __name__ == "__main__":
+ # [!Caution] The paths should be overrided for the local environment!
+ parser = argparse.ArgumentParser(description="Mapping files and paths")
+ parser.add_argument("--root", default=".", type=str)
+ args = parser.parse_args()
+
+ MAP_DIR = "./train_tools/data_utils/"
+
+ print("\n----------- Path Mapping for Labeled Data is Started... -----------\n")
+
+ map_labeled = os.path.join(MAP_DIR, "mapping_labeled.json")
+ map_dict = official_paths_labeled(args.root)
+ add_mapping_to_json(map_labeled, map_dict)
+
+ print("\n----------- Path Mapping for Tuning Data is Started... -----------\n")
+
+ map_labeled = os.path.join(MAP_DIR, "mapping_tuning.json")
+ map_dict = official_paths_tuning(args.root)
+ add_mapping_to_json(map_labeled, map_dict)
+
+ print("\n----------- Path Mapping for Public Data is Started... -----------\n")
+
+ map_public = os.path.join(MAP_DIR, "mapping_public.json")
+ map_dict = public_paths_labeled(args.root)
+ add_mapping_to_json(map_public, map_dict)
+
+ print("\n-------------- Path Mapping is Ended !!! ---------------------------\n")
diff --git a/annolid/segmentation/MEDIAR/image/examples/img2.tif b/annolid/segmentation/MEDIAR/image/examples/img2.tif
new file mode 100644
index 00000000..07780c71
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/examples/img2.tif differ
diff --git a/annolid/segmentation/MEDIAR/image/failure_cases.png b/annolid/segmentation/MEDIAR/image/failure_cases.png
new file mode 100644
index 00000000..fd9ab055
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/failure_cases.png differ
diff --git a/annolid/segmentation/MEDIAR/image/mediar_framework.png b/annolid/segmentation/MEDIAR/image/mediar_framework.png
new file mode 100644
index 00000000..8deb7a22
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/mediar_framework.png differ
diff --git a/annolid/segmentation/MEDIAR/image/mediar_model.PNG b/annolid/segmentation/MEDIAR/image/mediar_model.PNG
new file mode 100644
index 00000000..c842f9da
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/mediar_model.PNG differ
diff --git a/annolid/segmentation/MEDIAR/image/mediar_results.png b/annolid/segmentation/MEDIAR/image/mediar_results.png
new file mode 100644
index 00000000..8c05cf39
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/mediar_results.png differ
diff --git a/annolid/segmentation/MEDIAR/predict.py b/annolid/segmentation/MEDIAR/predict.py
new file mode 100644
index 00000000..02f86385
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/predict.py
@@ -0,0 +1,70 @@
+import torch
+import argparse, pprint
+
+from annolid.segmentation.MEDIAR.train_tools import *
+from annolid.segmentation.MEDIAR.SetupDict import MODELS, PREDICTOR
+
+# Set torch base print precision
+torch.set_printoptions(6)
+
+
+def main(args):
+ """Execute prediction and save the results"""
+
+ model_args = args.pred_setups.model
+ model = MODELS[model_args.name](**model_args.params)
+
+ if "ensemble" in args.pred_setups.name:
+ weights = torch.load(args.pred_setups.model_path1, map_location="cpu")
+ model.load_state_dict(weights, strict=False)
+
+ model_aux = MODELS[model_args.name](**model_args.params)
+ weights_aux = torch.load(args.pred_setups.model_path2, map_location="cpu")
+ model_aux.load_state_dict(weights_aux, strict=False)
+
+ predictor = PREDICTOR[args.pred_setups.name](
+ model,
+ model_aux,
+ args.pred_setups.device,
+ args.pred_setups.input_path,
+ args.pred_setups.output_path,
+ args.pred_setups.make_submission,
+ args.pred_setups.exp_name,
+ args.pred_setups.algo_params,
+ )
+
+ else:
+ weights = torch.load(args.pred_setups.model_path, map_location="cpu")
+ model.load_state_dict(weights, strict=False)
+
+ predictor = PREDICTOR[args.pred_setups.name](
+ model,
+ args.pred_setups.device,
+ args.pred_setups.input_path,
+ args.pred_setups.output_path,
+ args.pred_setups.make_submission,
+ args.pred_setups.exp_name,
+ args.pred_setups.algo_params,
+ )
+
+ _ = predictor.conduct_prediction()
+
+
+# Parser arguments for terminal execution
+parser = argparse.ArgumentParser(description="Config file processing")
+parser.add_argument(
+ "--config_path", default="./config/step3_prediction/base_prediction.json", type=str
+)
+args = parser.parse_args()
+
+#######################################################################################
+
+if __name__ == "__main__":
+ # Load configuration from .json file
+ opt = ConfLoader(args.config_path).opt
+
+ # Print configuration dictionary pretty
+ pprint_config(opt)
+
+ # Run experiment
+ main(opt)
diff --git a/annolid/segmentation/MEDIAR/predict_ensemble.py b/annolid/segmentation/MEDIAR/predict_ensemble.py
new file mode 100644
index 00000000..5edf47d6
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/predict_ensemble.py
@@ -0,0 +1,196 @@
+import os
+import torch
+import time
+from datetime import datetime
+import numpy as np
+from annolid.segmentation.MEDIAR.train_tools import *
+from annolid.segmentation.MEDIAR.train_tools.models import MEDIARFormer
+from annolid.utils.weights import WeightDownloader
+from annolid.segmentation.MEDIAR.core.MEDIAR import EnsemblePredictor
+from annolid.gui.shape import MaskShape
+from annolid.annotation.keypoints import save_labels
+from annolid.segmentation.MEDIAR.train_tools.data_utils.transforms import get_pred_transforms
+from annolid.utils.logger import logger
+
+
+class MEDIARPredictor(EnsemblePredictor):
+ """
+ Class for conducting cell detection using MEDIAR model.
+ https://github.com/Lee-Gihun/MEDIAR
+ @article{lee2022mediar,
+ title={Mediar: Harmony of data-centric and model-centric for multi-modality microscopy},
+ author={Lee, Gihun and Kim, SangMook and Kim, Joonkee and Yun, Se-Young},
+ journal={arXiv preprint arXiv:2212.03465},
+ year={2022}
+ }
+
+ Args:
+ weights_dir (str): Directory to store downloaded weights.
+ input_path (str): Directory containing input images.
+ output_path (str): Directory to store output predictions.
+ """
+
+ def __init__(self,
+ input_path="./data/images",
+ output_path="./output",
+ weights_dir="None",
+ ):
+ self.weights_dir = weights_dir
+ self.input_path = input_path
+ self.output_path = output_path
+ self.model1 = None
+ self.model2 = None
+ self.device = 'cuda' if torch.cuda.is_available() else 'cpu'
+ self.download_weights()
+ self.load_models()
+ super(MEDIARPredictor, self).__init__(self.model1, self.model2,
+ self.device, self.input_path,
+ self.output_path, algo_params={"use_tta": True})
+
+ def _setups(self):
+ self.pred_transforms = get_pred_transforms()
+ os.makedirs(self.output_path, exist_ok=True)
+
+ now = datetime.now()
+ dt_string = now.strftime("%m%d_%H%M")
+ self.exp_name = (
+ self.exp_name + dt_string if self.exp_name is not None else dt_string
+ )
+
+ self.img_names = [img_file for img_file in sorted(
+ os.listdir(self.input_path)) if '.json' not in img_file]
+ logger.info(f"Working on the images: {self.img_names}")
+
+ def download_weights(self, weights_dir=None):
+ """
+ Download pretrained weights for MEDIAR models.
+ """
+ if weights_dir is None:
+ self.weights_dir = os.path.join(
+ os.path.dirname(__file__), "weights")
+ downloader = WeightDownloader(self.weights_dir)
+
+ # Define weight URLs, expected checksums, and file names
+ weight_urls = [
+ "https://drive.google.com/uc?id=168MtudjTMLoq9YGTyoD2Rjl_d3Gy6c_L",
+ "https://drive.google.com/uc?id=1JJ2-QKTCk-G7sp5ddkqcifMxgnyOrXjx"
+ ]
+ expected_checksums = [
+ "e0ccb052828a9f05e21b2143939583c5",
+ "a595336926767afdf1ffb1baffd5ab7f"
+ ]
+ weight_file_names = ["from_phase1.pth", "from_phase2.pth"]
+
+ # Download weights for each URL
+ for url, checksum, file_name in zip(weight_urls, expected_checksums, weight_file_names):
+ downloader.download_weights(url, checksum, file_name)
+
+ def load_models(self):
+ """
+ Load pretrained MEDIAR models.
+ """
+ model_args = {
+ "classes": 3,
+ "decoder_channels": [1024, 512, 256, 128, 64],
+ "decoder_pab_channels": 256,
+ "encoder_name": 'mit_b5',
+ "in_channels": 3
+ }
+ self.model1 = MEDIARFormer(**model_args)
+ self.model1.load_state_dict(torch.load(f"{self.weights_dir}/from_phase1.pth",
+ map_location="cpu"), strict=False)
+
+ self.model2 = MEDIARFormer(**model_args)
+ self.model2.load_state_dict(torch.load(f"{self.weights_dir}/from_phase2.pth",
+ map_location="cpu"), strict=False)
+
+ @torch.no_grad()
+ def conduct_prediction(self):
+ self.model.to(self.device)
+ self.model.eval()
+ total_time = 0
+ total_times = []
+
+ for img_name in self.img_names:
+ img_data = self._get_img_data(img_name)
+ img_data = img_data.to(self.device)
+
+ start = time.time()
+
+ pred_mask = self._inference(img_data)
+ pred_mask = self._post_process(pred_mask.squeeze(0).cpu().numpy())
+
+ self.write_pred_mask(
+ pred_mask, self.output_path, img_name, self.make_submission
+ )
+ shape_list = self.save_prediction(
+ pred_mask, image_name=img_name)
+ end = time.time()
+
+ time_cost = end - start
+ total_times.append(time_cost)
+ total_time += time_cost
+ logger.info(
+ f"Prediction finished: {img_name}; img size = {img_data.shape}; costing: {time_cost:.2f}s"
+ )
+
+ logger.info(f"\n Total Time Cost: {total_time:.2f}s")
+
+ return shape_list
+
+ def _save_annotation(self, filename, mask_dict,
+ frame_shape,
+ img_ext='.png'):
+ if len(frame_shape) == 3:
+ height, width, _ = frame_shape
+ else:
+ height, width = frame_shape
+ label_list = []
+ for label_id, mask in mask_dict.items():
+ label = str(label_id)
+ current_shape = MaskShape(label=label,
+ flags={},
+ description='cell segmentation')
+ current_shape.mask = mask
+ _shapes = current_shape.toPolygons(
+ epsilon=2.0)
+ if len(_shapes) < 0:
+ continue
+ current_shape = _shapes[0]
+ points = [[point.x(), point.y()] for point in current_shape.points]
+ current_shape.points = points
+ label_list.append(current_shape)
+ img_abs_path = filename.replace('.json', img_ext)
+ save_labels(filename=filename,
+ imagePath=img_abs_path,
+ label_list=label_list,
+ height=height,
+ width=width,
+ save_image_to_json=False)
+ return label_list
+
+ def save_prediction(self, pred_mask, image_name="img1"):
+ """
+ Save prediction for a specific image.
+
+ Args:
+ image_name (str): Name of the input image file.
+ """
+ img_filename = os.path.join(self.input_path, image_name)
+ _, ext = os.path.splitext(img_filename)
+ # Replace the extension with ".json"
+ json_filename = img_filename.replace(ext, '.json')
+ json_filename = os.path.abspath(json_filename)
+ mask_dict = {f'cell_{label_id}': (pred_mask == label_id)
+ for label_id in np.unique(pred_mask)[1:]}
+ shape_list = self._save_annotation(json_filename, mask_dict,
+ pred_mask.shape, img_ext=ext)
+
+ cell_count = len(np.unique(pred_mask)) - 1 # exclude the background
+ logger.info(f"\n{cell_count} Cells detected!")
+ return shape_list
+
+
+if __name__ == "__main__":
+ mediar_predictor = MEDIARPredictor()
+ shape_list = mediar_predictor.conduct_prediction()
diff --git a/annolid/segmentation/MEDIAR/requirements.txt b/annolid/segmentation/MEDIAR/requirements.txt
new file mode 100644
index 00000000..7464efd9
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/requirements.txt
@@ -0,0 +1,14 @@
+fastremap==1.14.1
+monai==1.3.0
+numba==0.57.1
+numpy==1.24.3
+pandas==2.0.3
+pytz==2023.3.post1
+scipy==1.12.0
+segmentation_models_pytorch==0.3.3
+tifffile==2023.4.12
+torch==2.1.2
+tqdm==4.65.0
+wandb==0.16.2
+scikit-image
+matplotlib
diff --git a/annolid/segmentation/MEDIAR/train_tools/__init__.py b/annolid/segmentation/MEDIAR/train_tools/__init__.py
new file mode 100644
index 00000000..7614b006
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/__init__.py
@@ -0,0 +1,3 @@
+from .data_utils import *
+from .measures import *
+from .utils import *
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/__init__.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/__init__.py
new file mode 100644
index 00000000..e387c35e
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/__init__.py
@@ -0,0 +1 @@
+from .datasetter import *
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/CellAware.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/CellAware.py
new file mode 100644
index 00000000..24be12f6
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/CellAware.py
@@ -0,0 +1,88 @@
+import numpy as np
+import copy
+
+from monai.transforms import RandScaleIntensity, Compose
+from monai.transforms.compose import MapTransform
+from skimage.segmentation import find_boundaries
+
+
+__all__ = ["BoundaryExclusion", "IntensityDiversification"]
+
+
+class BoundaryExclusion(MapTransform):
+ """Map the cell boundary pixel labels to the background class (0)."""
+
+ def __init__(self, keys=["label"], allow_missing_keys=False):
+ super(BoundaryExclusion, self).__init__(keys, allow_missing_keys)
+
+ def __call__(self, data):
+ # Find and Exclude Boundary
+ label_original = data["label"]
+ label = copy.deepcopy(label_original)
+ boundary = find_boundaries(label, connectivity=1, mode="thick")
+ label[boundary] = 0
+
+ # Do not exclude if the cell is too small (< 14x14).
+ new_label = copy.deepcopy(label_original)
+ new_label[label == 0] = 0
+
+ cell_idx, cell_counts = np.unique(label_original, return_counts=True)
+
+ for k in range(len(cell_counts)):
+ if cell_counts[k] < 196:
+ new_label[label_original == cell_idx[k]] = cell_idx[k]
+
+ # Do not exclude if the pixels are at the image boundaries.
+ _, W, H = label_original.shape
+ bd = np.zeros_like(label_original, dtype=label.dtype)
+ bd[:, 2 : W - 2, 2 : H - 2] = 1
+ new_label += label_original * bd
+
+ # Assign the transformed label
+ data["label"] = new_label
+
+ return data
+
+
+class IntensityDiversification(MapTransform):
+ """Randomly rescale the intensity of cell pixels."""
+
+ def __init__(
+ self,
+ keys=["img"],
+ change_cell_ratio=0.4,
+ scale_factors=[0, 0.7],
+ allow_missing_keys=False,
+ ):
+ super(IntensityDiversification, self).__init__(keys, allow_missing_keys)
+
+ self.change_cell_ratio = change_cell_ratio
+ self.randscale_intensity = Compose(
+ [RandScaleIntensity(prob=1.0, factors=scale_factors)]
+ )
+
+ def __call__(self, data):
+ # Select cells to be transformed
+ cell_count = int(data["label"].max())
+ change_cell_count = int(cell_count * self.change_cell_ratio)
+ change_cell = np.random.choice(cell_count, change_cell_count, replace=False)
+
+ mask = copy.deepcopy(data["label"])
+
+ for i in range(cell_count):
+ cell_id = i + 1
+
+ if cell_id not in change_cell:
+ mask[mask == cell_id] = 0
+
+ mask[mask > 0] = 1
+
+ # Conduct intensity transformation for the selected cells
+ img_original = copy.deepcopy((1 - mask) * data["img"])
+ img_transformed = copy.deepcopy(mask * data["img"])
+ img_transformed = self.randscale_intensity(img_transformed)
+
+ # Assign the transformed image
+ data["img"] = img_original + img_transformed
+
+ return data
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/LoadImage.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/LoadImage.py
new file mode 100644
index 00000000..aad77b13
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/LoadImage.py
@@ -0,0 +1,161 @@
+import numpy as np
+import tifffile as tif
+import skimage.io as io
+from typing import Optional, Sequence, Union
+from monai.config import DtypeLike, PathLike, KeysCollection
+from monai.utils import ensure_tuple
+from monai.data.utils import is_supported_format, optional_import, ensure_tuple_rep
+from monai.data.image_reader import ImageReader, NumpyReader
+from monai.transforms import LoadImage, LoadImaged
+from monai.utils.enums import PostFix
+
+DEFAULT_POST_FIX = PostFix.meta()
+itk, has_itk = optional_import("itk", allow_namespace_pkg=True)
+
+__all__ = [
+ "CustomLoadImaged",
+ "CustomLoadImageD",
+ "CustomLoadImageDict",
+ "CustomLoadImage",
+]
+
+
+class CustomLoadImage(LoadImage):
+ """
+ Load image file or files from provided path based on reader.
+ If reader is not specified, this class automatically chooses readers
+ based on the supported suffixes and in the following order:
+
+ - User-specified reader at runtime when calling this loader.
+ - User-specified reader in the constructor of `LoadImage`.
+ - Readers from the last to the first in the registered list.
+ - Current default readers: (nii, nii.gz -> NibabelReader), (png, jpg, bmp -> PILReader),
+ (npz, npy -> NumpyReader), (nrrd -> NrrdReader), (DICOM file -> ITKReader).
+
+ [!Caution] This overriding replaces the original ITK with Custom UnifiedITKReader.
+ """
+
+ def __init__(
+ self,
+ reader=None,
+ image_only: bool = False,
+ dtype: DtypeLike = np.float32,
+ ensure_channel_first: bool = False,
+ *args,
+ **kwargs,
+ ) -> None:
+ super(CustomLoadImage, self).__init__(
+ reader, image_only, dtype, ensure_channel_first, *args, **kwargs
+ )
+
+ # Adding TIFFReader. Although ITK Reader supports ".tiff" files, sometimes fails to load images.
+ self.readers = []
+ self.register(UnifiedITKReader(*args, **kwargs))
+
+
+class CustomLoadImaged(LoadImaged):
+ """
+ Dictionary-based wrapper of `CustomLoadImage`.
+ """
+
+ def __init__(
+ self,
+ keys: KeysCollection,
+ reader: Optional[Union[ImageReader, str]] = None,
+ dtype: DtypeLike = np.float32,
+ meta_keys: Optional[KeysCollection] = None,
+ meta_key_postfix: str = DEFAULT_POST_FIX,
+ overwriting: bool = False,
+ image_only: bool = False,
+ ensure_channel_first: bool = False,
+ simple_keys=False,
+ allow_missing_keys: bool = False,
+ *args,
+ **kwargs,
+ ) -> None:
+ super(CustomLoadImaged, self).__init__(
+ keys,
+ reader,
+ dtype,
+ meta_keys,
+ meta_key_postfix,
+ overwriting,
+ image_only,
+ ensure_channel_first,
+ simple_keys,
+ allow_missing_keys,
+ *args,
+ **kwargs,
+ )
+
+ # Assign CustomLoader
+ self._loader = CustomLoadImage(
+ reader, image_only, dtype, ensure_channel_first, *args, **kwargs
+ )
+ if not isinstance(meta_key_postfix, str):
+ raise TypeError(
+ f"meta_key_postfix must be a str but is {type(meta_key_postfix).__name__}."
+ )
+ self.meta_keys = (
+ ensure_tuple_rep(None, len(self.keys))
+ if meta_keys is None
+ else ensure_tuple(meta_keys)
+ )
+ if len(self.keys) != len(self.meta_keys):
+ raise ValueError("meta_keys should have the same length as keys.")
+ self.meta_key_postfix = ensure_tuple_rep(meta_key_postfix, len(self.keys))
+ self.overwriting = overwriting
+
+
+class UnifiedITKReader(NumpyReader):
+ """
+ Unified Reader to read ".tif" and ".tiff files".
+ As the tifffile reads the images as numpy arrays, it inherits from the NumpyReader.
+ """
+
+ def __init__(
+ self, channel_dim: Optional[int] = None, **kwargs,
+ ):
+ super(UnifiedITKReader, self).__init__(channel_dim=channel_dim, **kwargs)
+ self.kwargs = kwargs
+ self.channel_dim = channel_dim
+
+ def verify_suffix(self, filename: Union[Sequence[PathLike], PathLike]) -> bool:
+ """Verify whether the file format is supported by TIFF Reader."""
+
+ suffixes: Sequence[str] = ["tif", "tiff", "png", "jpg", "bmp", "jpeg",]
+ return has_itk or is_supported_format(filename, suffixes)
+
+ def read(self, data: Union[Sequence[PathLike], PathLike], **kwargs):
+ """Read Images from the file."""
+ img_ = []
+
+ filenames: Sequence[PathLike] = ensure_tuple(data)
+ kwargs_ = self.kwargs.copy()
+ kwargs_.update(kwargs)
+
+ for name in filenames:
+ name = f"{name}"
+
+ if name.endswith(".tif") or name.endswith(".tiff"):
+ _obj = tif.imread(name)
+ else:
+ try:
+ _obj = itk.imread(name, **kwargs_)
+ _obj = itk.array_view_from_image(_obj, keep_axes=False)
+ except:
+ _obj = io.imread(name)
+
+ if len(_obj.shape) == 2:
+ _obj = np.repeat(np.expand_dims(_obj, axis=-1), 3, axis=-1)
+ elif len(_obj.shape) == 3 and _obj.shape[-1] > 3:
+ _obj = _obj[:, :, :3]
+ else:
+ pass
+
+ img_.append(_obj)
+
+ return img_ if len(filenames) > 1 else img_[0]
+
+
+CustomLoadImageD = CustomLoadImageDict = CustomLoadImaged
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/NormalizeImage.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/NormalizeImage.py
new file mode 100644
index 00000000..08a21509
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/NormalizeImage.py
@@ -0,0 +1,77 @@
+import numpy as np
+from skimage import exposure
+from monai.config import KeysCollection
+
+from monai.transforms.transform import Transform
+from monai.transforms.compose import MapTransform
+
+from typing import Dict, Hashable, Mapping
+
+
+__all__ = [
+ "CustomNormalizeImage",
+ "CustomNormalizeImageD",
+ "CustomNormalizeImageDict",
+ "CustomNormalizeImaged",
+]
+
+
+class CustomNormalizeImage(Transform):
+ """Normalize the image."""
+
+ def __init__(self, percentiles=[0, 99.5], channel_wise=False):
+ self.lower, self.upper = percentiles
+ self.channel_wise = channel_wise
+
+ def _normalize(self, img) -> np.ndarray:
+ non_zero_vals = img[np.nonzero(img)]
+ percentiles = np.percentile(non_zero_vals, [self.lower, self.upper])
+ img_norm = exposure.rescale_intensity(
+ img, in_range=(percentiles[0], percentiles[1]), out_range="uint8"
+ )
+
+ return img_norm.astype(np.uint8)
+
+ def __call__(self, img: np.ndarray) -> np.ndarray:
+ if self.channel_wise:
+ pre_img_data = np.zeros(img.shape, dtype=np.uint8)
+ for i in range(img.shape[-1]):
+ img_channel_i = img[:, :, i]
+
+ if len(img_channel_i[np.nonzero(img_channel_i)]) > 0:
+ pre_img_data[:, :, i] = self._normalize(img_channel_i)
+
+ img = pre_img_data
+
+ else:
+ img = self._normalize(img)
+
+ return img
+
+
+class CustomNormalizeImaged(MapTransform):
+ """Dictionary-based wrapper of NormalizeImage"""
+
+ def __init__(
+ self,
+ keys: KeysCollection,
+ percentiles=[1, 99],
+ channel_wise: bool = False,
+ allow_missing_keys: bool = False,
+ ):
+ super(CustomNormalizeImageD, self).__init__(keys, allow_missing_keys)
+ self.normalizer = CustomNormalizeImage(percentiles, channel_wise)
+
+ def __call__(
+ self, data: Mapping[Hashable, np.ndarray]
+ ) -> Dict[Hashable, np.ndarray]:
+
+ d = dict(data)
+
+ for key in self.keys:
+ d[key] = self.normalizer(d[key])
+
+ return d
+
+
+CustomNormalizeImageD = CustomNormalizeImageDict = CustomNormalizeImaged
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/__init__.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/__init__.py
new file mode 100644
index 00000000..33fc9ebe
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/__init__.py
@@ -0,0 +1,3 @@
+from .LoadImage import *
+from .NormalizeImage import *
+from .CellAware import *
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/modalities.pkl b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/modalities.pkl
new file mode 100644
index 00000000..4f867966
Binary files /dev/null and b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/modalities.pkl differ
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/datasetter.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/datasetter.py
new file mode 100644
index 00000000..6a6b3317
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/datasetter.py
@@ -0,0 +1,208 @@
+from torch.utils.data import DataLoader
+from monai.data import Dataset
+import pickle
+
+from .transforms import (
+ train_transforms,
+ public_transforms,
+ valid_transforms,
+ tuning_transforms,
+ unlabeled_transforms,
+)
+from .utils import split_train_valid, path_decoder
+
+DATA_LABEL_DICT_PICKLE_FILE = "./train_tools/data_utils/custom/modalities.pkl"
+
+__all__ = [
+ "get_dataloaders_labeled",
+ "get_dataloaders_public",
+ "get_dataloaders_unlabeled",
+]
+
+
+def get_dataloaders_labeled(
+ root,
+ mapping_file,
+ mapping_file_tuning,
+ join_mapping_file=None,
+ valid_portion=0.0,
+ batch_size=8,
+ amplified=False,
+ relabel=False,
+):
+ """Set DataLoaders for labeled datasets.
+
+ Args:
+ root (str): root directory
+ mapping_file (str): json file for mapping dataset
+ valid_portion (float, optional): portion of valid datasets. Defaults to 0.1.
+ batch_size (int, optional): batch size. Defaults to 8.
+ shuffle (bool, optional): shuffles dataloader. Defaults to True.
+ num_workers (int, optional): number of workers for each datalaoder. Defaults to 5.
+
+ Returns:
+ dict: dictionary of data loaders.
+ """
+
+ # Get list of data dictionaries from decoded paths
+ data_dicts = path_decoder(root, mapping_file)
+ tuning_dicts = path_decoder(root, mapping_file_tuning, no_label=True)
+
+ if amplified:
+ with open(DATA_LABEL_DICT_PICKLE_FILE, "rb") as f:
+ data_label_dict = pickle.load(f)
+
+ data_point_dict = {}
+
+ for label, data_lst in data_label_dict.items():
+ data_point_dict[label] = []
+
+ for d_idx in data_lst:
+ try:
+ data_point_dict[label].append(data_dicts[d_idx])
+ except:
+ print(label, d_idx)
+
+ data_dicts = []
+
+ for label, data_points in data_point_dict.items():
+ len_data_points = len(data_points)
+
+ if len_data_points >= 50:
+ data_dicts += data_points
+ else:
+ for i in range(50):
+ data_dicts.append(data_points[i % len_data_points])
+
+ data_transforms = train_transforms
+
+ if join_mapping_file is not None:
+ data_dicts += path_decoder(root, join_mapping_file)
+ data_transforms = public_transforms
+
+ if relabel:
+ for elem in data_dicts:
+ cell_idx = int(elem["label"].split("_label.tiff")[0].split("_")[-1])
+ if cell_idx in range(340, 499):
+ new_label = elem["label"].replace(
+ "/data/CellSeg/Official/Train_Labeled/labels/",
+ "/CellSeg/pretrained_train_ext/",
+ )
+ elem["label"] = new_label
+
+ # Split datasets as Train/Valid
+ train_dicts, valid_dicts = split_train_valid(
+ data_dicts, valid_portion=valid_portion
+ )
+
+ # Obtain datasets with transforms
+ trainset = Dataset(train_dicts, transform=data_transforms)
+ validset = Dataset(valid_dicts, transform=valid_transforms)
+ tuningset = Dataset(tuning_dicts, transform=tuning_transforms)
+
+ # Set dataloader for Trainset
+ train_loader = DataLoader(
+ trainset, batch_size=batch_size, shuffle=True, num_workers=5
+ )
+
+ # Set dataloader for Validset (Batch size is fixed as 1)
+ valid_loader = DataLoader(validset, batch_size=1, shuffle=False,)
+
+ # Set dataloader for Tuningset (Batch size is fixed as 1)
+ tuning_loader = DataLoader(tuningset, batch_size=1, shuffle=False)
+
+ # Form dataloaders as dictionary
+ dataloaders = {
+ "train": train_loader,
+ "valid": valid_loader,
+ "tuning": tuning_loader,
+ }
+
+ return dataloaders
+
+
+def get_dataloaders_public(
+ root, mapping_file, valid_portion=0.0, batch_size=8,
+):
+ """Set DataLoaders for labeled datasets.
+
+ Args:
+ root (str): root directory
+ mapping_file (str): json file for mapping dataset
+ valid_portion (float, optional): portion of valid datasets. Defaults to 0.1.
+ batch_size (int, optional): batch size. Defaults to 8.
+ shuffle (bool, optional): shuffles dataloader. Defaults to True.
+
+ Returns:
+ dict: dictionary of data loaders.
+ """
+
+ # Get list of data dictionaries from decoded paths
+ data_dicts = path_decoder(root, mapping_file)
+
+ # Split datasets as Train/Valid
+ train_dicts, _ = split_train_valid(data_dicts, valid_portion=valid_portion)
+
+ trainset = Dataset(train_dicts, transform=public_transforms)
+ # Set dataloader for Trainset
+ train_loader = DataLoader(
+ trainset, batch_size=batch_size, shuffle=True, num_workers=5
+ )
+
+ # Form dataloaders as dictionary
+ dataloaders = {
+ "public": train_loader,
+ }
+
+ return dataloaders
+
+
+def get_dataloaders_unlabeled(
+ root, mapping_file, batch_size=8, shuffle=True, num_workers=5,
+):
+ """Set dataloaders for unlabeled dataset."""
+ # Get list of data dictionaries from decoded paths
+ unlabeled_dicts = path_decoder(root, mapping_file, no_label=True, unlabeled=True)
+
+ # Obtain datasets with transforms
+ unlabeled_dicts, _ = split_train_valid(unlabeled_dicts, valid_portion=0)
+ unlabeled_set = Dataset(unlabeled_dicts, transform=unlabeled_transforms)
+
+ # Set dataloader for Unlabeled dataset
+ unlabeled_loader = DataLoader(
+ unlabeled_set, batch_size=batch_size, shuffle=shuffle, num_workers=num_workers
+ )
+
+ dataloaders = {
+ "unlabeled": unlabeled_loader,
+ }
+
+ return dataloaders
+
+
+def get_dataloaders_unlabeled_psuedo(
+ root, mapping_file, batch_size=8, shuffle=True, num_workers=5,
+):
+
+ # Get list of data dictionaries from decoded paths
+ unlabeled_psuedo_dicts = path_decoder(
+ root, mapping_file, no_label=False, unlabeled=True
+ )
+
+ # Obtain datasets with transforms
+ unlabeled_psuedo_dicts, _ = split_train_valid(
+ unlabeled_psuedo_dicts, valid_portion=0
+ )
+ unlabeled_psuedo_set = Dataset(unlabeled_psuedo_dicts, transform=train_transforms)
+
+ # Set dataloader for Unlabeled dataset
+ unlabeled_psuedo_loader = DataLoader(
+ unlabeled_psuedo_set,
+ batch_size=batch_size,
+ shuffle=shuffle,
+ num_workers=num_workers,
+ )
+
+ dataloaders = {"unlabeled": unlabeled_psuedo_loader}
+
+ return dataloaders
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/transforms.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/transforms.py
new file mode 100644
index 00000000..00560027
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/transforms.py
@@ -0,0 +1,148 @@
+from .custom import *
+
+from monai.transforms import *
+
+__all__ = [
+ "train_transforms",
+ "public_transforms",
+ "valid_transforms",
+ "tuning_transforms",
+ "unlabeled_transforms",
+]
+
+train_transforms = Compose(
+ [
+ # >>> Load and refine data --- img: (H, W, 3); label: (H, W)
+ CustomLoadImaged(keys=["img", "label"], image_only=True),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img", "label"], channel_dim=-1),
+ RemoveRepeatedChanneld(keys=["label"], repeats=3), # label: (H, W)
+ ScaleIntensityd(keys=["img"], allow_missing_keys=True), # Do not scale label
+ # >>> Spatial transforms
+ RandZoomd(
+ keys=["img", "label"],
+ prob=0.5,
+ min_zoom=0.25,
+ max_zoom=1.5,
+ mode=["area", "nearest"],
+ keep_size=False,
+ ),
+ SpatialPadd(keys=["img", "label"], spatial_size=512),
+ RandSpatialCropd(keys=["img", "label"], roi_size=512, random_size=False),
+ RandAxisFlipd(keys=["img", "label"], prob=0.5),
+ RandRotate90d(keys=["img", "label"], prob=0.5, spatial_axes=[0, 1]),
+ IntensityDiversification(keys=["img", "label"], allow_missing_keys=True),
+ # # >>> Intensity transforms
+ RandGaussianNoised(keys=["img"], prob=0.25, mean=0, std=0.1),
+ RandAdjustContrastd(keys=["img"], prob=0.25, gamma=(1, 2)),
+ RandGaussianSmoothd(keys=["img"], prob=0.25, sigma_x=(1, 2)),
+ RandHistogramShiftd(keys=["img"], prob=0.25, num_control_points=3),
+ RandGaussianSharpend(keys=["img"], prob=0.25),
+ EnsureTyped(keys=["img", "label"]),
+ ]
+)
+
+
+public_transforms = Compose(
+ [
+ CustomLoadImaged(keys=["img", "label"], image_only=True),
+ BoundaryExclusion(keys=["label"]),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img", "label"], channel_dim=-1),
+ RemoveRepeatedChanneld(keys=["label"], repeats=3), # label: (H, W)
+ ScaleIntensityd(keys=["img"], allow_missing_keys=True), # Do not scale label
+ # >>> Spatial transforms
+ SpatialPadd(keys=["img", "label"], spatial_size=512),
+ RandSpatialCropd(keys=["img", "label"], roi_size=512, random_size=False),
+ RandAxisFlipd(keys=["img", "label"], prob=0.5),
+ RandRotate90d(keys=["img", "label"], prob=0.5, spatial_axes=[0, 1]),
+ Rotate90d(k=1, keys=["label"], spatial_axes=(0, 1)),
+ Flipd(keys=["label"], spatial_axis=0),
+ EnsureTyped(keys=["img", "label"]),
+ ]
+)
+
+
+valid_transforms = Compose(
+ [
+ CustomLoadImaged(keys=["img", "label"], allow_missing_keys=True, image_only=True),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img", "label"], allow_missing_keys=True, channel_dim=-1),
+ RemoveRepeatedChanneld(keys=["label"], repeats=3),
+ ScaleIntensityd(keys=["img"], allow_missing_keys=True),
+ EnsureTyped(keys=["img", "label"], allow_missing_keys=True),
+ ]
+)
+
+tuning_transforms = Compose(
+ [
+ CustomLoadImaged(keys=["img"], image_only=True),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img"], channel_dim=-1),
+ ScaleIntensityd(keys=["img"]),
+ EnsureTyped(keys=["img"]),
+ ]
+)
+
+unlabeled_transforms = Compose(
+ [
+ # >>> Load and refine data --- img: (H, W, 3); label: (H, W)
+ CustomLoadImaged(keys=["img"], image_only=True),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img"], channel_dim=-1),
+ RandZoomd(
+ keys=["img"],
+ prob=0.5,
+ min_zoom=0.25,
+ max_zoom=1.25,
+ mode=["area"],
+ keep_size=False,
+ ),
+ ScaleIntensityd(keys=["img"], allow_missing_keys=True), # Do not scale label
+ # >>> Spatial transforms
+ SpatialPadd(keys=["img"], spatial_size=512),
+ RandSpatialCropd(keys=["img"], roi_size=512, random_size=False),
+ EnsureTyped(keys=["img"]),
+ ]
+)
+
+
+def get_pred_transforms():
+ """Prediction preprocessing"""
+ pred_transforms = Compose(
+ [
+ # >>> Load and refine data
+ CustomLoadImage(image_only=True),
+ CustomNormalizeImage(channel_wise=False, percentiles=[0.0, 99.5]),
+ EnsureChannelFirst(channel_dim=-1), # image: (3, H, W)
+ ScaleIntensity(),
+ EnsureType(data_type="tensor"),
+ ]
+ )
+
+ return pred_transforms
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/utils.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/utils.py
new file mode 100644
index 00000000..6e926a3c
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/utils.py
@@ -0,0 +1,84 @@
+import os
+import json
+import numpy as np
+
+__all__ = ["split_train_valid", "path_decoder"]
+
+
+def split_train_valid(data_dicts, valid_portion=0.1):
+ """Split train/validata data according to the given proportion"""
+
+ train_dicts, valid_dicts = data_dicts, []
+ if valid_portion > 0:
+
+ # Obtain & shuffle data indices
+ num_data_dicts = len(data_dicts)
+ indices = np.arange(num_data_dicts)
+ np.random.shuffle(indices)
+
+ # Divide train/valid indices by the proportion
+ valid_size = int(num_data_dicts * valid_portion)
+ train_indices = indices[valid_size:]
+ valid_indices = indices[:valid_size]
+
+ # Assign data dicts by split indices
+ train_dicts = [data_dicts[idx] for idx in train_indices]
+ valid_dicts = [data_dicts[idx] for idx in valid_indices]
+
+ print(
+ "\n(DataLoaded) Training data size: %d, Validation data size: %d\n"
+ % (len(train_dicts), len(valid_dicts))
+ )
+
+ return train_dicts, valid_dicts
+
+
+def path_decoder(root, mapping_file, no_label=False, unlabeled=False):
+ """Decode img/label file paths from root & mapping directory.
+
+ Args:
+ root (str):
+ mapping_file (str): json file containing image & label file paths.
+ no_label (bool, optional): whether to include "label" key. Defaults to False.
+
+ Returns:
+ list: list of dictionary. (ex. [{"img": img_path, "label": label_path}, ...])
+ """
+
+ data_dicts = []
+
+ with open(mapping_file, "r") as file:
+ data = json.load(file)
+
+ for map_key in data.keys():
+
+ # If no_label, assign "img" key only
+ if no_label:
+ data_dict_item = [
+ {"img": os.path.join(root, elem["img"]),} for elem in data[map_key]
+ ]
+
+ # If label exists, assign both "img" and "label" keys
+ else:
+ data_dict_item = [
+ {
+ "img": os.path.join(root, elem["img"]),
+ "label": os.path.join(root, elem["label"]),
+ }
+ for elem in data[map_key]
+ ]
+
+ # Add refined datasets to be returned
+ data_dicts += data_dict_item
+
+ if unlabeled:
+ refined_data_dicts = []
+
+ # Exclude the corrupted image to prevent errror
+ for data_dict in data_dicts:
+ if "00504" not in data_dict["img"]:
+ refined_data_dicts.append(data_dict)
+
+ data_dicts = refined_data_dicts
+
+ return data_dicts
diff --git a/annolid/segmentation/MEDIAR/train_tools/measures.py b/annolid/segmentation/MEDIAR/train_tools/measures.py
new file mode 100644
index 00000000..b17509a4
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/measures.py
@@ -0,0 +1,200 @@
+"""
+Adapted from the following references:
+[1] https://github.com/JunMa11/NeurIPS-CellSeg/blob/main/baseline/compute_metric.py
+[2] https://github.com/stardist/stardist/blob/master/stardist/matching.py
+
+"""
+
+import numpy as np
+from skimage import segmentation
+from scipy.optimize import linear_sum_assignment
+from numba import jit
+
+__all__ = ["evaluate_f1_score_cellseg", "evaluate_f1_score"]
+
+
+def evaluate_f1_score_cellseg(masks_true, masks_pred, threshold=0.5):
+ """
+ Get confusion elements for cell segmentation results.
+ Boundary pixels are not considered during evaluation.
+ """
+
+ if np.prod(masks_true.shape) < (5000 * 5000):
+ masks_true = _remove_boundary_cells(masks_true.astype(np.int32))
+ masks_pred = _remove_boundary_cells(masks_pred.astype(np.int32))
+
+ tp, fp, fn = get_confusion(masks_true, masks_pred, threshold)
+
+ # Compute by Patch-based way for large images
+ else:
+ H, W = masks_true.shape
+ roi_size = 2000
+
+ # Get patch grid by roi_size
+ if H % roi_size != 0:
+ n_H = H // roi_size + 1
+ new_H = roi_size * n_H
+ else:
+ n_H = H // roi_size
+ new_H = H
+
+ if W % roi_size != 0:
+ n_W = W // roi_size + 1
+ new_W = roi_size * n_W
+ else:
+ n_W = W // roi_size
+ new_W = W
+
+ # Allocate values on the grid
+ gt_pad = np.zeros((new_H, new_W), dtype=masks_true.dtype)
+ pred_pad = np.zeros((new_H, new_W), dtype=masks_true.dtype)
+ gt_pad[:H, :W] = masks_true
+ pred_pad[:H, :W] = masks_pred
+
+ tp, fp, fn = 0, 0, 0
+
+ # Calculate confusion elements for each patch
+ for i in range(n_H):
+ for j in range(n_W):
+ gt_roi = _remove_boundary_cells(
+ gt_pad[
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ]
+ )
+ pred_roi = _remove_boundary_cells(
+ pred_pad[
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ]
+ )
+ tp_i, fp_i, fn_i = get_confusion(gt_roi, pred_roi, threshold)
+ tp += tp_i
+ fp += fp_i
+ fn += fn_i
+
+ # Calculate f1 score
+ precision, recall, f1_score = evaluate_f1_score(tp, fp, fn)
+
+ return precision, recall, f1_score
+
+
+def evaluate_f1_score(tp, fp, fn):
+ """Evaluate F1-score for the given confusion elements"""
+
+ # Do not Compute on trivial results
+ if tp == 0:
+ precision, recall, f1_score = 0, 0, 0
+
+ else:
+ precision = tp / (tp + fp)
+ recall = tp / (tp + fn)
+ f1_score = 2 * (precision * recall) / (precision + recall)
+
+ return precision, recall, f1_score
+
+
+def _remove_boundary_cells(mask):
+ """Remove cells on the boundary from the mask"""
+
+ # Identify boundary cells
+ W, H = mask.shape
+ bd = np.ones((W, H))
+ bd[2 : W - 2, 2 : H - 2] = 0
+ bd_cells = np.unique(mask * bd)
+
+ # Remove cells on the boundary
+ for i in bd_cells[1:]:
+ mask[mask == i] = 0
+
+ # Allocate labels as sequential manner
+ new_label, _, _ = segmentation.relabel_sequential(mask)
+
+ return new_label
+
+
+def get_confusion(masks_true, masks_pred, threshold=0.5):
+ """Calculate confusion matrix elements: (TP, FP, FN)"""
+ num_gt_instances = np.max(masks_true)
+ num_pred_instances = np.max(masks_pred)
+
+ if num_pred_instances == 0:
+ print("No segmentation results!")
+ tp, fp, fn = 0, 0, 0
+
+ else:
+ # Calculate IoU and exclude background label (0)
+ iou = _get_iou(masks_true, masks_pred)
+ iou = iou[1:, 1:]
+
+ # Calculate true positives
+ tp = _get_true_positive(iou, threshold)
+ fp = num_pred_instances - tp
+ fn = num_gt_instances - tp
+
+ return tp, fp, fn
+
+
+def _get_true_positive(iou, threshold=0.5):
+ """Get true positive (TP) pixels at the given threshold"""
+
+ # Number of instances to be matched
+ num_matched = min(iou.shape[0], iou.shape[1])
+
+ # Find optimal matching by using IoU as tie-breaker
+ costs = -(iou >= threshold).astype(np.float) - iou / (2 * num_matched)
+ matched_gt_label, matched_pred_label = linear_sum_assignment(costs)
+
+ # Consider as the same instance only if the IoU is above the threshold
+ match_ok = iou[matched_gt_label, matched_pred_label] >= threshold
+ tp = match_ok.sum()
+
+ return tp
+
+
+def _get_iou(masks_true, masks_pred):
+ """Get the iou between masks_true and masks_pred"""
+
+ # Get overlap matrix (GT Instances Num, Pred Instance Num)
+ overlap = _label_overlap(masks_true, masks_pred)
+
+ # Predicted instance pixels
+ n_pixels_pred = np.sum(overlap, axis=0, keepdims=True)
+
+ # GT instance pixels
+ n_pixels_true = np.sum(overlap, axis=1, keepdims=True)
+
+ # Calculate intersection of union (IoU)
+ union = n_pixels_pred + n_pixels_true - overlap
+ iou = overlap / union
+
+ # Ensure numerical values
+ iou[np.isnan(iou)] = 0.0
+
+ return iou
+
+
+@jit(nopython=True)
+def _label_overlap(x, y):
+ """Get pixel overlaps between two masks
+
+ Parameters
+ ------------
+ x, y (np array; dtype int): 0=NO masks; 1,2... are mask labels
+
+ Returns
+ ------------
+ overlap (np array; dtype int): Overlaps of size [x.max()+1, y.max()+1]
+ """
+
+ # Make as 1D array
+ x, y = x.ravel(), y.ravel()
+
+ # Preallocate a Contact Map matrix
+ overlap = np.zeros((1 + x.max(), 1 + y.max()), dtype=np.uint)
+
+ # Calculate the number of shared pixels for each label
+ for i in range(len(x)):
+ overlap[x[i], y[i]] += 1
+
+ return overlap
diff --git a/annolid/segmentation/MEDIAR/train_tools/models/MEDIARFormer.py b/annolid/segmentation/MEDIAR/train_tools/models/MEDIARFormer.py
new file mode 100644
index 00000000..f7f19f2b
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/models/MEDIARFormer.py
@@ -0,0 +1,102 @@
+import torch
+import torch.nn as nn
+
+from segmentation_models_pytorch import MAnet
+from segmentation_models_pytorch.base.modules import Activation
+
+__all__ = ["MEDIARFormer"]
+
+
+class MEDIARFormer(MAnet):
+ """MEDIAR-Former Model"""
+
+ def __init__(
+ self,
+ encoder_name="mit_b5", # Default encoder
+ encoder_weights="imagenet", # Pre-trained weights
+ decoder_channels=(1024, 512, 256, 128, 64), # Decoder configuration
+ decoder_pab_channels=256, # Decoder Pyramid Attention Block channels
+ in_channels=3, # Number of input channels
+ classes=3, # Number of output classes
+ ):
+ # Initialize the MAnet model with provided parameters
+ super().__init__(
+ encoder_name=encoder_name,
+ encoder_weights=encoder_weights,
+ decoder_channels=decoder_channels,
+ decoder_pab_channels=decoder_pab_channels,
+ in_channels=in_channels,
+ classes=classes,
+ )
+
+ # Remove the default segmentation head as it's not used in this architecture
+ self.segmentation_head = None
+
+ # Modify all activation functions in the encoder and decoder from ReLU to Mish
+ _convert_activations(self.encoder, nn.ReLU, nn.Mish(inplace=True))
+ _convert_activations(self.decoder, nn.ReLU, nn.Mish(inplace=True))
+
+ # Add custom segmentation heads for different segmentation tasks
+ self.cellprob_head = DeepSegmentationHead(
+ in_channels=decoder_channels[-1], out_channels=1
+ )
+ self.gradflow_head = DeepSegmentationHead(
+ in_channels=decoder_channels[-1], out_channels=2
+ )
+
+ def forward(self, x):
+ """Forward pass through the network"""
+ # Ensure the input shape is correct
+ self.check_input_shape(x)
+
+ # Encode the input and then decode it
+ features = self.encoder(x)
+ decoder_output = self.decoder(*features)
+
+ # Generate masks for cell probability and gradient flows
+ cellprob_mask = self.cellprob_head(decoder_output)
+ gradflow_mask = self.gradflow_head(decoder_output)
+
+ # Concatenate the masks for output
+ masks = torch.cat([gradflow_mask, cellprob_mask], dim=1)
+
+ return masks
+
+
+class DeepSegmentationHead(nn.Sequential):
+ """Custom segmentation head for generating specific masks"""
+
+ def __init__(
+ self, in_channels, out_channels, kernel_size=3, activation=None, upsampling=1
+ ):
+ # Define a sequence of layers for the segmentation head
+ layers = [
+ nn.Conv2d(
+ in_channels,
+ in_channels // 2,
+ kernel_size=kernel_size,
+ padding=kernel_size // 2,
+ ),
+ nn.Mish(inplace=True),
+ nn.BatchNorm2d(in_channels // 2),
+ nn.Conv2d(
+ in_channels // 2,
+ out_channels,
+ kernel_size=kernel_size,
+ padding=kernel_size // 2,
+ ),
+ nn.UpsamplingBilinear2d(scale_factor=upsampling)
+ if upsampling > 1
+ else nn.Identity(),
+ Activation(activation) if activation else nn.Identity(),
+ ]
+ super().__init__(*layers)
+
+
+def _convert_activations(module, from_activation, to_activation):
+ """Recursively convert activation functions in a module"""
+ for name, child in module.named_children():
+ if isinstance(child, from_activation):
+ setattr(module, name, to_activation)
+ else:
+ _convert_activations(child, from_activation, to_activation)
diff --git a/annolid/segmentation/MEDIAR/train_tools/models/__init__.py b/annolid/segmentation/MEDIAR/train_tools/models/__init__.py
new file mode 100644
index 00000000..cc8bcdb6
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/models/__init__.py
@@ -0,0 +1 @@
+from .MEDIARFormer import *
diff --git a/annolid/segmentation/MEDIAR/train_tools/utils.py b/annolid/segmentation/MEDIAR/train_tools/utils.py
new file mode 100644
index 00000000..73b704df
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/utils.py
@@ -0,0 +1,70 @@
+import torch
+import numpy as np
+import os, json, random
+from pprint import pprint
+
+__all__ = ["ConfLoader", "directory_setter", "random_seeder", "pprint_config"]
+
+
+class ConfLoader:
+ """
+ Load json config file using DictWithAttributeAccess object_hook.
+ ConfLoader(conf_name).opt attribute is the result of loading json config file.
+ """
+
+ class DictWithAttributeAccess(dict):
+ """
+ This inner class makes dict to be accessed same as class attribute.
+ For example, you can use opt.key instead of the opt['key'].
+ """
+
+ def __getattr__(self, key):
+ return self[key]
+
+ def __setattr__(self, key, value):
+ self[key] = value
+
+ def __init__(self, conf_name):
+ self.conf_name = conf_name
+ self.opt = self.__get_opt()
+
+ def __load_conf(self):
+ with open(self.conf_name, "r") as conf:
+ opt = json.load(
+ conf, object_hook=lambda dict: self.DictWithAttributeAccess(dict)
+ )
+ return opt
+
+ def __get_opt(self):
+ opt = self.__load_conf()
+ opt = self.DictWithAttributeAccess(opt)
+
+ return opt
+
+
+def directory_setter(path="./results", make_dir=False):
+ """
+ Make dictionary if not exists.
+ """
+ if not os.path.exists(path) and make_dir:
+ os.makedirs(path) # make dir if not exist
+ print("directory %s is created" % path)
+
+ if not os.path.isdir(path):
+ raise NotADirectoryError(
+ "%s is not valid. set make_dir=True to make dir." % path
+ )
+
+
+def random_seeder(seed):
+ """Fix randomness."""
+ torch.manual_seed(seed)
+ np.random.seed(seed)
+ random.seed(seed)
+ torch.backends.cudnn.deterministic = True
+ torch.backends.cudnn.benchmark = False
+
+def pprint_config(opt):
+ print("\n" + "=" * 50 + " Configuration " + "=" * 50)
+ pprint(opt, compact=True)
+ print("=" * 115 + "\n")
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/viz.py b/annolid/segmentation/MEDIAR/viz.py
new file mode 100644
index 00000000..7b39fe43
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/viz.py
@@ -0,0 +1,29 @@
+import os
+from skimage import io
+import numpy as np
+
+def show_and_count_cells(output_path, img_name="img2_label.tiff", cmap="cividis"):
+ pred_path = os.path.join(output_path, img_name)
+
+ try:
+ pred = io.imread(pred_path)
+ except Exception as e:
+ print(f"Error loading image: {e}")
+ return
+
+ if pred is None:
+ print("Image is empty or could not be loaded.")
+ return
+
+ try:
+ io.imshow(pred, cmap=cmap)
+ io.show()
+ except Exception as e:
+ print(f"Error displaying image: {e}")
+
+ cell_count = len(np.unique(pred)) - 1 # Exclude the background
+ print(f"\n{cell_count} Cells detected!")
+
+# Example usage:
+output_path = "./results/mediar_base_prediction"
+show_and_count_cells(output_path,'OpenTest_004_label.tiff')
\ No newline at end of file
diff --git a/setup.py b/setup.py
index 0b32bb99..9f23c651 100644
--- a/setup.py
+++ b/setup.py
@@ -47,6 +47,13 @@
'Pillow>=9.3.0,<=9.5.0',
"chardet>=5.2.0",
"scikit-learn-extra >= 0.3.0",
+ 'monai>=1.3.0',
+ 'segmentation-models-pytorch>=0.3.3',
+ 'fastremap>=1.14.1',
+ 'numba>=0.57.1',
+ 'tifffile>=2023.4.12',
+ 'wandb>=0.16.2',
+ 'scikit-image',
# "segment-anything @ git+https://github.com/facebookresearch/segment-anything.git",
"segment-anything @ git+https://github.com/SysCV/sam-hq.git",
],
+
+### **Test Dataset**
+
+
+
+
+# 5. Reproducing
+
+### **Our Environment**
+| Computing Infrastructure| |
+|-------------------------|----------------------------------------------------------------------|
+| System | Ubuntu 18.04.5 LTS |
+| CPU | AMD EPYC 7543 32-Core Processor CPU@2.26GHz |
+| RAM | 500GB; 3.125MT/s |
+| GPU (number and type) | NVIDIA A5000 (24GB) 2ea |
+| CUDA version | 11.7 |
+| Programming language | Python 3.9 |
+| Deep learning framework | Pytorch (v1.12, with torchvision v0.13.1) |
+| Code dependencies | MONAI (v0.9.0), Segmentation Models (v0.3.0) |
+| Specific dependencies | None |
+
+To install requirements:
+
+```
+pip install -r requirements.txt
+wandb off
+```
+
+## Dataset
+- The datasets directories under the root should the following structure:
+
+```
+ Root
+ ├── Datasets
+ │ ├── images (images can have various extensions: .tif, .tiff, .png, .bmp ...)
+ │ │ ├── cell_00001.png
+ │ │ ├── cell_00002.tif
+ │ │ ├── cell_00003.xxx
+ │ │ ├── ...
+ │ └── labels (labels must have .tiff extension.)
+ │ │ ├── cell_00001_label.tiff
+ │ │ ├── cell_00002.label.tiff
+ │ │ ├── cell_00003.label.tiff
+ │ │ ├── ...
+ └── ...
+```
+
+Before execute the codes, run the follwing code to generate path mappting json file:
+
+```python
+python ./generate_mapping.py --root=
+```
+
+## Training
+
+To train the model(s) in the paper, run the following command:
+
+```python
+python ./main.py --config_path=
+```
+Configuration files are in `./config/*`. We provide the pretraining, fine-tuning, and prediction configs. You can refer to the configuration options in the `./config/mediar_example.json`. We also implemented the official challenge baseline code in our framework. You can run the baseline code by running the `./config/baseline.json`.
+
+## Inference
+
+To conduct prediction on the testing cases, run the following command:
+
+```python
+python predict.py --config_path=
+```
+
+## Evaluation
+If you have the labels run the following command for evaluation:
+
+```python
+python ./evaluate.py --pred_path= --gt_path=
+```
+
+The configuration files for `predict.py` is slightly different. Please refer to the config files in `./config/step3_prediction/*`.
+## Trained Models
+
+You can download MEDIAR pretrained and finetuned models here:
+
+- [Google Drive Link](https://drive.google.com/drive/folders/1RgMxHIT7WsKNjir3wXSl7BrzlpS05S18?usp=share_link).
+
+## Citation of this Work
+```
+@article{lee2022mediar,
+ title={Mediar: Harmony of data-centric and model-centric for multi-modality microscopy},
+ author={Lee, Gihun and Kim, SangMook and Kim, Joonkee and Yun, Se-Young},
+ journal={arXiv preprint arXiv:2212.03465},
+ year={2022}
+}
+```
+
diff --git a/annolid/segmentation/MEDIAR/SetupDict.py b/annolid/segmentation/MEDIAR/SetupDict.py
new file mode 100644
index 00000000..d9b7aebd
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/SetupDict.py
@@ -0,0 +1,39 @@
+import torch.optim as optim
+import torch.optim.lr_scheduler as lr_scheduler
+import monai
+
+from annolid.segmentation.MEDIAR import core
+from annolid.segmentation.MEDIAR.train_tools import models
+from annolid.segmentation.MEDIAR.train_tools.models import *
+
+__all__ = ["TRAINER", "OPTIMIZER", "SCHEDULER"]
+
+TRAINER = {
+ "baseline": core.Baseline.Trainer,
+ "mediar": core.MEDIAR.Trainer,
+}
+
+PREDICTOR = {
+ "baseline": core.Baseline.Predictor,
+ "mediar": core.MEDIAR.Predictor,
+ "ensemble_mediar": core.MEDIAR.EnsemblePredictor,
+}
+
+MODELS = {
+ "unet": monai.networks.nets.UNet,
+ "unetr": monai.networks.nets.unetr.UNETR,
+ "swinunetr": monai.networks.nets.SwinUNETR,
+ "mediar-former": models.MEDIARFormer,
+}
+
+OPTIMIZER = {
+ "sgd": optim.SGD,
+ "adam": optim.Adam,
+ "adamw": optim.AdamW,
+}
+
+SCHEDULER = {
+ "step": lr_scheduler.StepLR,
+ "multistep": lr_scheduler.MultiStepLR,
+ "cosine": lr_scheduler.CosineAnnealingLR,
+}
diff --git a/annolid/segmentation/MEDIAR/__init__.py b/annolid/segmentation/MEDIAR/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/annolid/segmentation/MEDIAR/config/baseline.json b/annolid/segmentation/MEDIAR/config/baseline.json
new file mode 100644
index 00000000..186910a4
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/baseline.json
@@ -0,0 +1,60 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/data/CellSeg/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "tuning_mapping_file": "/home/gihun/CellSeg/train_tools/data_utils/mapping_tuning.json",
+ "batch_size": 8,
+ "valid_portion": 0.1
+ },
+ "unlabeled":{
+ "enabled": false
+ },
+ "public":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "swinunetr",
+ "params": {
+ "img_size": 512,
+ "in_channels": 3,
+ "out_channels": 3,
+ "spatial_dims": 2
+ },
+ "pretrained":{
+ "enabled": false
+ }
+ },
+ "trainer": {
+ "name": "baseline",
+ "params": {
+ "num_epochs": 200,
+ "valid_frequency": 1,
+ "device": "cuda:0",
+ "algo_params": {}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "params": {"lr": 5e-5}
+ },
+ "scheduler":{
+ "enabled": false
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/data/CellSeg/Official/TuningSet",
+ "output_path": "./results/baseline",
+ "make_submission": true,
+ "exp_name": "baseline",
+ "algo_params": {}
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Baseline",
+ "name": "baseline"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/mediar_example.json b/annolid/segmentation/MEDIAR/config/mediar_example.json
new file mode 100644
index 00000000..11c99495
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/mediar_example.json
@@ -0,0 +1,72 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/data/CellSeg/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "amplified": false,
+ "batch_size": 8,
+ "valid_portion": 0.1
+ },
+ "public":{
+ "enabled": true,
+ "params":{
+ "root": "/home/gihun/data/CellSeg/",
+ "mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "batch_size": 1
+ }
+ },
+ "unlabeled":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {
+ "encoder_name": "mit_b5",
+ "encoder_weights": "imagenet",
+ "decoder_channels": [1024, 512, 256, 128, 64],
+ "decoder_pab_channels": 256,
+ "in_channels": 3,
+ "classes": 3
+ },
+ "pretrained":{
+ "enabled": false,
+ "weights": "./weights/pretrained/phase2.pth",
+ "strict": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 200,
+ "valid_frequency": 1,
+ "device": "cuda:0",
+ "amp": true,
+ "algo_params": {"with_public": false}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "params": {"lr": 5e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 100, "eta_min": 1e-7}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/data/CellSeg/Official/TuningSet",
+ "output_path": "./mediar_example",
+ "make_submission": true,
+ "exp_name": "mediar_example",
+ "algo_params": {"use_tta": false}
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "MEDIAR",
+ "name": "mediar_example"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/pred/pred_mediar.json b/annolid/segmentation/MEDIAR/config/pred/pred_mediar.json
new file mode 100644
index 00000000..65493771
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/pred/pred_mediar.json
@@ -0,0 +1,17 @@
+{
+ "pred_setups":{
+ "name": "medair",
+ "input_path":"input_path",
+ "output_path": "./test",
+ "make_submission": true,
+ "model_path": "model_path",
+ "device": "cuda:0",
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": false
+ }
+ }
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step1_pretraining/phase1.json b/annolid/segmentation/MEDIAR/config/step1_pretraining/phase1.json
new file mode 100644
index 00000000..a8baee70
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step1_pretraining/phase1.json
@@ -0,0 +1,55 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "mapping_file_tuning": "/home/gihun/MEDIAR/train_tools/data_utils/mapping_tuning.json",
+ "batch_size": 9,
+ "valid_portion": 0
+ },
+ "public":{
+ "enabled": false,
+ "params":{}
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 80,
+ "valid_frequency": 10,
+ "device": "cuda:0",
+ "amp": true,
+ "algo_params": {"with_public": false}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "ft_rate": 1.0,
+ "params": {"lr": 5e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 80, "eta_min": 1e-6}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./mediar_pretrained_phase1",
+ "make_submission": false
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Pretraining",
+ "name": "phase1"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step1_pretraining/phase2.json b/annolid/segmentation/MEDIAR/config/step1_pretraining/phase2.json
new file mode 100644
index 00000000..b13e4bb4
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step1_pretraining/phase2.json
@@ -0,0 +1,58 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "mapping_file_tuning": "/home/gihun/MEDIAR/train_tools/data_utils/mapping_tuning.json",
+ "join_mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "batch_size": 9,
+ "valid_portion": 0
+ },
+ "unlabeled":{
+ "enabled": false
+ },
+ "public":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 60,
+ "valid_frequency": 10,
+ "device": "cuda:0",
+ "amp": true,
+ "algo_params": {"with_public": false}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "ft_rate": 1.0,
+ "params": {"lr": 5e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 60, "eta_min": 1e-6}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./mediar_pretrain_phase2",
+ "make_submission": false
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Pretraining",
+ "name": "phase2"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning1.json b/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning1.json
new file mode 100644
index 00000000..dbdea432
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning1.json
@@ -0,0 +1,66 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "mapping_file_tuning": "/home/gihun/MEDIAR/train_tools/data_utils/mapping_tuning.json",
+ "amplified": true,
+ "batch_size": 8,
+ "valid_portion": 0.0
+ },
+ "public":{
+ "enabled": false,
+ "params":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "batch_size": 1
+ }
+ },
+ "unlabeled":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": true,
+ "weights": "./weights/pretrained/phase1.pth",
+ "strict": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 200,
+ "valid_frequency": 1,
+ "device": "cuda:7",
+ "amp": true,
+ "algo_params": {"with_public": false}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "params": {"lr": 2e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 100, "eta_min": 1e-7}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./results/",
+ "make_submission": true,
+ "exp_name": "mediar_from_phase1",
+ "algo_params": {"use_tta": false}
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Fine-tuning",
+ "name": "from_phase1"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning2.json b/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning2.json
new file mode 100644
index 00000000..149a0f5d
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step2_finetuning/finetuning2.json
@@ -0,0 +1,66 @@
+{
+ "data_setups":{
+ "labeled":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_labeled.json",
+ "mapping_file_tuning": "/home/gihun/MEDIAR/train_tools/data_utils/mapping_tuning.json",
+ "amplified": true,
+ "batch_size": 8,
+ "valid_portion": 0.0
+ },
+ "public":{
+ "enabled": true,
+ "params":{
+ "root": "/home/gihun/MEDIAR/",
+ "mapping_file": "./train_tools/data_utils/mapping_public.json",
+ "batch_size": 1
+ }
+ },
+ "unlabeled":{
+ "enabled": false
+ }
+ },
+ "train_setups":{
+ "model":{
+ "name": "mediar-former",
+ "params": {},
+ "pretrained":{
+ "enabled": true,
+ "weights": "./weights/pretrained/phase2.pth",
+ "strict": false
+ }
+ },
+ "trainer": {
+ "name": "mediar",
+ "params": {
+ "num_epochs": 50,
+ "valid_frequency": 1,
+ "device": "cuda:0",
+ "amp": true,
+ "algo_params": {"with_public": true}
+ }
+ },
+ "optimizer":{
+ "name": "adamw",
+ "params": {"lr": 2e-5}
+ },
+ "scheduler":{
+ "enabled": true,
+ "name": "cosine",
+ "params": {"T_max": 100, "eta_min": 1e-7}
+ },
+ "seed": 19940817
+ },
+ "pred_setups":{
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./results/from_phase2",
+ "make_submission": true,
+ "exp_name": "mediar_from_phase2",
+ "algo_params": {"use_tta": false}
+ },
+ "wandb_setups":{
+ "project": "CellSeg",
+ "group": "Fine-tuning",
+ "name": "from_phase2"
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step3_prediction/base_prediction.json b/annolid/segmentation/MEDIAR/config/step3_prediction/base_prediction.json
new file mode 100644
index 00000000..e253daea
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step3_prediction/base_prediction.json
@@ -0,0 +1,16 @@
+{
+ "pred_setups":{
+ "name": "mediar",
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./results/mediar_base_prediction",
+ "make_submission": true,
+ "model_path": "./weights/finetuned/from_phase1.pth",
+ "device": "cuda:7",
+ "model":{
+ "name": "mediar-former",
+ "params": {}
+ },
+ "exp_name": "mediar_p1_base",
+ "algo_params": {"use_tta": false}
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/config/step3_prediction/ensemble_tta.json b/annolid/segmentation/MEDIAR/config/step3_prediction/ensemble_tta.json
new file mode 100644
index 00000000..4d3910b6
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/config/step3_prediction/ensemble_tta.json
@@ -0,0 +1,23 @@
+{
+ "pred_setups":{
+ "name": "ensemble_mediar",
+ "input_path":"/home/gihun/MEDIAR/data/Official/Tuning/images",
+ "output_path": "./results/mediar_ensemble_tta",
+ "make_submission": true,
+ "model_path1": "./weights/finetuned/from_phase1.pth",
+ "model_path2": "./weights/finetuned/from_phase2.pth",
+ "device": "cuda:0",
+ "model":{
+ "name": "mediar-former",
+ "params": {
+ "encoder_name":"mit_b5",
+ "decoder_channels": [1024, 512, 256, 128, 64],
+ "decoder_pab_channels": 256,
+ "in_channels":3,
+ "classes":3
+ }
+ },
+ "exp_name": "mediar_ensemble_tta",
+ "algo_params": {"use_tta": true}
+ }
+}
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/core/BasePredictor.py b/annolid/segmentation/MEDIAR/core/BasePredictor.py
new file mode 100644
index 00000000..c1c9753d
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/BasePredictor.py
@@ -0,0 +1,126 @@
+import torch
+import numpy as np
+import time
+import os
+import tifffile as tif
+
+from datetime import datetime
+from zipfile import ZipFile
+from pytz import timezone
+from annolid.segmentation.MEDIAR.train_tools.data_utils.transforms import get_pred_transforms
+
+
+class BasePredictor:
+ def __init__(
+ self,
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission=False,
+ exp_name=None,
+ algo_params=None,
+ ):
+ self.model = model
+ self.device = device
+ self.input_path = input_path
+ self.output_path = output_path
+ self.make_submission = make_submission
+ self.exp_name = exp_name
+
+ # Assign algoritm-specific arguments
+ if algo_params:
+ self.__dict__.update((k, v) for k, v in algo_params.items())
+
+ # Prepare inference environments
+ self._setups()
+
+ @torch.no_grad()
+ def conduct_prediction(self):
+ self.model.to(self.device)
+ self.model.eval()
+ total_time = 0
+ total_times = []
+
+ for img_name in self.img_names:
+ img_data = self._get_img_data(img_name)
+ img_data = img_data.to(self.device)
+
+ start = time.time()
+
+ pred_mask = self._inference(img_data)
+ pred_mask = self._post_process(pred_mask.squeeze(0).cpu().numpy())
+
+ self.write_pred_mask(
+ pred_mask, self.output_path, img_name, self.make_submission
+ )
+ self.save_prediction(
+ pred_mask, image_name=img_name)
+ end = time.time()
+
+ time_cost = end - start
+ total_times.append(time_cost)
+ total_time += time_cost
+ print(
+ f"Prediction finished: {img_name}; img size = {img_data.shape}; costing: {time_cost:.2f}s"
+ )
+
+ print(f"\n Total Time Cost: {total_time:.2f}s")
+
+ if self.make_submission:
+ fname = "%s.zip" % self.exp_name
+
+ os.makedirs("./submissions", exist_ok=True)
+ submission_path = os.path.join("./submissions", fname)
+
+ with ZipFile(submission_path, "w") as zipObj2:
+ pred_names = sorted(os.listdir(self.output_path))
+ for pred_name in pred_names:
+ pred_path = os.path.join(self.output_path, pred_name)
+ zipObj2.write(pred_path)
+
+ print("\n>>>>> Submission file is saved at: %s\n" % submission_path)
+
+ return time_cost
+
+ def write_pred_mask(self, pred_mask,
+ output_dir,
+ image_name,
+ submission=False):
+
+ # All images should contain at least 5 cells
+ if submission:
+ if not (np.max(pred_mask) > 5):
+ print("[!Caution] Only %d Cells Detected!!!\n" %
+ np.max(pred_mask))
+
+ file_name = image_name.split(".")[0]
+ file_name = file_name + "_label.tiff"
+ file_path = os.path.join(output_dir, file_name)
+
+ tif.imwrite(file_path, pred_mask, compression="zlib")
+
+ def _setups(self):
+ self.pred_transforms = get_pred_transforms()
+ os.makedirs(self.output_path, exist_ok=True)
+
+ now = datetime.now(timezone("Asia/Seoul"))
+ dt_string = now.strftime("%m%d_%H%M")
+ self.exp_name = (
+ self.exp_name + dt_string if self.exp_name is not None else dt_string
+ )
+
+ self.img_names = sorted(os.listdir(self.input_path))
+
+ def _get_img_data(self, img_name):
+ img_path = os.path.join(self.input_path, img_name)
+ img_data = self.pred_transforms(img_path)
+ img_data = img_data.unsqueeze(0)
+
+ return img_data
+
+ def _inference(self, img_data):
+ raise NotImplementedError
+
+ def _post_process(self, pred_mask):
+ raise NotImplementedError
diff --git a/annolid/segmentation/MEDIAR/core/BaseTrainer.py b/annolid/segmentation/MEDIAR/core/BaseTrainer.py
new file mode 100644
index 00000000..102f1b4e
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/BaseTrainer.py
@@ -0,0 +1,240 @@
+import torch
+import numpy as np
+from tqdm import tqdm
+from monai.inferers import sliding_window_inference
+from monai.metrics import CumulativeAverage
+from monai.transforms import (
+ Activations,
+ AsDiscrete,
+ Compose,
+ EnsureType,
+)
+
+import os, sys
+import copy
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../")))
+
+from annolid.segmentation.MEDIAR.core.utils import print_learning_device, print_with_logging
+from annolid.segmentation.MEDIAR.train_tools.measures import evaluate_f1_score_cellseg
+
+
+class BaseTrainer:
+ """Abstract base class for trainer implementations"""
+
+ def __init__(
+ self,
+ model,
+ dataloaders,
+ optimizer,
+ scheduler=None,
+ criterion=None,
+ num_epochs=100,
+ device="cuda:0",
+ no_valid=False,
+ valid_frequency=1,
+ amp=False,
+ algo_params=None,
+ ):
+ self.model = model.to(device)
+ self.dataloaders = dataloaders
+ self.optimizer = optimizer
+ self.scheduler = scheduler
+ self.criterion = criterion
+ self.num_epochs = num_epochs
+ self.no_valid = no_valid
+ self.valid_frequency = valid_frequency
+ self.device = device
+ self.amp = amp
+ self.best_weights = None
+ self.best_f1_score = 0.1
+
+ # FP-16 Scaler
+ self.scaler = torch.cuda.amp.GradScaler() if amp else None
+
+ # Assign algoritm-specific arguments
+ if algo_params:
+ self.__dict__.update((k, v) for k, v in algo_params.items())
+
+ # Cumulitive statistics
+ self.loss_metric = CumulativeAverage()
+ self.f1_metric = CumulativeAverage()
+
+ # Post-processing functions
+ self.post_pred = Compose(
+ [EnsureType(), Activations(softmax=True), AsDiscrete(threshold=0.5)]
+ )
+ self.post_gt = Compose([EnsureType(), AsDiscrete(to_onehot=None)])
+
+ def train(self):
+ """Train the model"""
+
+ # Print learning device name
+ print_learning_device(self.device)
+
+ # Learning process
+ for epoch in range(1, self.num_epochs + 1):
+ print(f"[Round {epoch}/{self.num_epochs}]")
+
+ # Train Epoch Phase
+ print(">>> Train Epoch")
+ train_results = self._epoch_phase("train")
+ print_with_logging(train_results, epoch)
+
+ if self.scheduler is not None:
+ self.scheduler.step()
+
+ if epoch % self.valid_frequency == 0:
+ if not self.no_valid:
+ # Valid Epoch Phase
+ print(">>> Valid Epoch")
+ valid_results = self._epoch_phase("valid")
+ print_with_logging(valid_results, epoch)
+
+ if "Valid_F1_Score" in valid_results.keys():
+ current_f1_score = valid_results["Valid_F1_Score"]
+ self._update_best_model(current_f1_score)
+ else:
+ print(">>> TuningSet Epoch")
+ tuning_cell_counts = self._tuningset_evaluation()
+ tuning_count_dict = {"TuningSet_Cell_Count": tuning_cell_counts}
+ print_with_logging(tuning_count_dict, epoch)
+
+ current_cell_count = tuning_cell_counts
+ self._update_best_model(current_cell_count)
+
+ print("-" * 50)
+
+ self.best_f1_score = 0
+
+ if self.best_weights is not None:
+ self.model.load_state_dict(self.best_weights)
+
+ def _epoch_phase(self, phase):
+ """Learning process for 1 Epoch (for different phases).
+
+ Args:
+ phase (str): "train", "valid", "test"
+
+ Returns:
+ dict: statistics for the phase results
+ """
+ phase_results = {}
+
+ # Set model mode
+ self.model.train() if phase == "train" else self.model.eval()
+
+ # Epoch process
+ for batch_data in tqdm(self.dataloaders[phase]):
+ images = batch_data["img"].to(self.device)
+ labels = batch_data["label"].to(self.device)
+ self.optimizer.zero_grad()
+
+ # Forward pass
+ with torch.set_grad_enabled(phase == "train"):
+ outputs = self.model(images)
+ loss = self.criterion(outputs, labels)
+ self.loss_metric.append(loss)
+
+ # Backward pass
+ if phase == "train":
+ loss.backward()
+ self.optimizer.step()
+
+ # Update metrics
+ phase_results = self._update_results(
+ phase_results, self.loss_metric, "loss", phase
+ )
+
+ return phase_results
+
+ @torch.no_grad()
+ def _tuningset_evaluation(self):
+ cell_counts_total = []
+ self.model.eval()
+
+ for batch_data in tqdm(self.dataloaders["tuning"]):
+ images = batch_data["img"].to(self.device)
+ if images.shape[-1] > 5000:
+ continue
+
+ outputs = sliding_window_inference(
+ images,
+ roi_size=512,
+ sw_batch_size=4,
+ predictor=self.model,
+ padding_mode="constant",
+ mode="gaussian",
+ )
+
+ outputs = outputs.squeeze(0)
+ outputs, _ = self._post_process(outputs, None)
+ count = len(np.unique(outputs) - 1)
+ cell_counts_total.append(count)
+
+ cell_counts_total_sum = np.sum(cell_counts_total)
+ print("Cell Counts Total: (%d)" % (cell_counts_total_sum))
+
+ return cell_counts_total_sum
+
+ def _update_results(self, phase_results, metric, metric_key, phase="train"):
+ """Aggregate and flush metrics
+
+ Args:
+ phase_results (dict): base dictionary to log metrics
+ metric (_type_): cumulated metrics
+ metric_key (_type_): name of metric
+ phase (str, optional): current phase name. Defaults to "train".
+
+ Returns:
+ dict: dictionary of metrics for the current phase
+ """
+
+ # Refine metrics name
+ metric_key = "_".join([phase, metric_key]).title()
+
+ # Aggregate metrics
+ metric_item = round(metric.aggregate().item(), 4)
+
+ # Log metrics to dictionary
+ phase_results[metric_key] = metric_item
+
+ # Flush metrics
+ metric.reset()
+
+ return phase_results
+
+ def _update_best_model(self, current_f1_score):
+ if current_f1_score > self.best_f1_score:
+ self.best_weights = copy.deepcopy(self.model.state_dict())
+ self.best_f1_score = current_f1_score
+ print(
+ "\n>>>> Update Best Model with score: {}\n".format(self.best_f1_score)
+ )
+ else:
+ pass
+
+ def _inference(self, images, phase="train"):
+ """inference methods for different phase"""
+ if phase != "train":
+ outputs = sliding_window_inference(
+ images,
+ roi_size=512,
+ sw_batch_size=4,
+ predictor=self.model,
+ padding_mode="reflect",
+ mode="gaussian",
+ overlap=0.5,
+ )
+ else:
+ outputs = self.model(images)
+
+ return outputs
+
+ def _post_process(self, outputs, labels):
+ return outputs, labels
+
+ def _get_f1_metric(self, masks_pred, masks_true):
+ f1_score = evaluate_f1_score_cellseg(masks_true, masks_pred)[-1]
+
+ return f1_score
diff --git a/annolid/segmentation/MEDIAR/core/Baseline/Predictor.py b/annolid/segmentation/MEDIAR/core/Baseline/Predictor.py
new file mode 100644
index 00000000..b4036f40
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/Baseline/Predictor.py
@@ -0,0 +1,59 @@
+import torch
+import os, sys
+from skimage import morphology, measure
+from monai.inferers import sliding_window_inference
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+from annolid.segmentation.MEDIAR.core.BasePredictor import BasePredictor
+
+__all__ = ["Predictor"]
+
+
+class Predictor(BasePredictor):
+ def __init__(
+ self,
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission=False,
+ exp_name=None,
+ algo_params=None,
+ ):
+ super(Predictor, self).__init__(
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission,
+ exp_name,
+ algo_params,
+ )
+
+ def _inference(self, img_data):
+ pred_mask = sliding_window_inference(
+ img_data,
+ 512,
+ 4,
+ self.model,
+ padding_mode="constant",
+ mode="gaussian",
+ overlap=0.6,
+ )
+
+ return pred_mask
+
+ def _post_process(self, pred_mask):
+ # Get probability map from the predicted logits
+ pred_mask = torch.from_numpy(pred_mask)
+ pred_mask = torch.softmax(pred_mask, dim=0)
+ pred_mask = pred_mask[1].cpu().numpy()
+
+ # Apply morphological post-processing
+ pred_mask = pred_mask > 0.5
+ pred_mask = morphology.remove_small_holes(pred_mask, connectivity=1)
+ pred_mask = morphology.remove_small_objects(pred_mask, 16)
+ pred_mask = measure.label(pred_mask)
+
+ return pred_mask
diff --git a/annolid/segmentation/MEDIAR/core/Baseline/Trainer.py b/annolid/segmentation/MEDIAR/core/Baseline/Trainer.py
new file mode 100644
index 00000000..1c4585e1
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/Baseline/Trainer.py
@@ -0,0 +1,115 @@
+from tqdm import tqdm
+from annolid.segmentation.MEDIAR.train_tools.measures import evaluate_f1_score_cellseg
+from annolid.segmentation.MEDIAR.core.Baseline.utils import create_interior_onehot, identify_instances_from_classmap
+from annolid.segmentation.MEDIAR.core.BaseTrainer import BaseTrainer
+import torch
+import os
+import sys
+import monai
+
+from monai.data import decollate_batch
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+
+__all__ = ["Trainer"]
+
+
+class Trainer(BaseTrainer):
+ def __init__(
+ self,
+ model,
+ dataloaders,
+ optimizer,
+ scheduler=None,
+ criterion=None,
+ num_epochs=100,
+ device="cuda:0",
+ no_valid=False,
+ valid_frequency=1,
+ amp=False,
+ algo_params=None,
+ ):
+ super(Trainer, self).__init__(
+ model,
+ dataloaders,
+ optimizer,
+ scheduler,
+ criterion,
+ num_epochs,
+ device,
+ no_valid,
+ valid_frequency,
+ amp,
+ algo_params,
+ )
+
+ # Dice loss as segmentation criterion
+ self.criterion = monai.losses.DiceCELoss(softmax=True)
+
+ def _epoch_phase(self, phase):
+ """Learning process for 1 Epoch."""
+
+ phase_results = {}
+
+ # Set model mode
+ self.model.train() if phase == "train" else self.model.eval()
+
+ # Epoch process
+ for batch_data in tqdm(self.dataloaders[phase]):
+ images = batch_data["img"].to(self.device)
+ labels = batch_data["label"].to(self.device)
+ self.optimizer.zero_grad()
+
+ # Map label masks to 3-class onehot map
+ labels_onehot = create_interior_onehot(labels)
+
+ # Forward pass
+ with torch.set_grad_enabled(phase == "train"):
+ outputs = self._inference(images, phase)
+ loss = self.criterion(outputs, labels_onehot)
+ self.loss_metric.append(loss)
+
+ if phase != "train":
+ f1_score = self._get_f1_metric(outputs, labels)
+ self.f1_metric.append(f1_score)
+
+ # Backward pass
+ if phase == "train":
+ # For the mixed precision training
+ if self.amp:
+ self.scaler.scale(loss).backward()
+ self.scaler.unscale_(self.optimizer)
+ self.scaler.step(self.optimizer)
+ self.scaler.update()
+
+ else:
+ loss.backward()
+ self.optimizer.step()
+
+ # Update metrics
+ phase_results = self._update_results(
+ phase_results, self.loss_metric, "loss", phase
+ )
+
+ if phase != "train":
+ phase_results = self._update_results(
+ phase_results, self.f1_metric, "f1_score", phase
+ )
+
+ return phase_results
+
+ def _post_process(self, outputs, labels_onehot):
+ """Conduct post-processing for outputs & labels."""
+ outputs = [self.post_pred(i) for i in decollate_batch(outputs)]
+ labels_onehot = [self.post_gt(i)
+ for i in decollate_batch(labels_onehot)]
+
+ return outputs, labels_onehot
+
+ def _get_f1_metric(self, masks_pred, masks_true):
+ masks_pred = identify_instances_from_classmap(masks_pred[0])
+ masks_true = masks_true.squeeze(0).squeeze(0).cpu().numpy()
+ f1_score = evaluate_f1_score_cellseg(masks_true, masks_pred)[-1]
+
+ return f1_score
diff --git a/annolid/segmentation/MEDIAR/core/Baseline/__init__.py b/annolid/segmentation/MEDIAR/core/Baseline/__init__.py
new file mode 100644
index 00000000..5d102375
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/Baseline/__init__.py
@@ -0,0 +1,2 @@
+from .Trainer import *
+from .Predictor import *
diff --git a/annolid/segmentation/MEDIAR/core/Baseline/utils.py b/annolid/segmentation/MEDIAR/core/Baseline/utils.py
new file mode 100644
index 00000000..4fd559de
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/Baseline/utils.py
@@ -0,0 +1,80 @@
+"""
+Adapted from the following references:
+[1] https://github.com/JunMa11/NeurIPS-CellSeg/blob/main/baseline/model_training_3class.py
+
+"""
+
+import torch
+import numpy as np
+from skimage import segmentation, morphology, measure
+import monai
+
+
+__all__ = ["create_interior_onehot", "identify_instances_from_classmap"]
+
+
+@torch.no_grad()
+def identify_instances_from_classmap(
+ class_map, cell_class=1, threshold=0.5, from_logits=True
+):
+ """Identification of cell instances from the class map"""
+
+ if from_logits:
+ class_map = torch.softmax(class_map, dim=0) # (C, H, W)
+
+ # Convert probability map to binary mask
+ pred_mask = class_map[cell_class].cpu().numpy()
+
+ # Apply morphological postprocessing
+ pred_mask = pred_mask > threshold
+ pred_mask = morphology.remove_small_holes(pred_mask, connectivity=1)
+ pred_mask = morphology.remove_small_objects(pred_mask, 16)
+ pred_mask = measure.label(pred_mask)
+
+ return pred_mask
+
+
+@torch.no_grad()
+def create_interior_onehot(inst_maps):
+ """
+ interior : (H,W), np.uint8
+ three-class map, values: 0,1,2
+ 0: background
+ 1: interior
+ 2: boundary
+ """
+ device = inst_maps.device
+
+ # Get (np.int16) array corresponding to label masks: (B, 1, H, W)
+ inst_maps = inst_maps.squeeze(1).cpu().numpy().astype(np.int16)
+
+ interior_maps = []
+
+ for inst_map in inst_maps:
+ # Create interior-edge map
+ boundary = segmentation.find_boundaries(inst_map, mode="inner")
+
+ # Refine interior-edge map
+ boundary = morphology.binary_dilation(boundary, morphology.disk(1))
+
+ # Assign label classes
+ interior_temp = np.logical_and(~boundary, inst_map > 0)
+
+ # interior_temp[boundary] = 0
+ interior_temp = morphology.remove_small_objects(interior_temp, min_size=16)
+ interior = np.zeros_like(inst_map, dtype=np.uint8)
+ interior[interior_temp] = 1
+ interior[boundary] = 2
+
+ interior_maps.append(interior)
+
+ # Aggregate interior_maps for batch
+ interior_maps = np.stack(interior_maps, axis=0).astype(np.uint8)
+
+ # Reshape as original label shape: (B, H, W)
+ interior_maps = torch.from_numpy(interior_maps).unsqueeze(1).to(device)
+
+ # Obtain one-hot map for batch
+ interior_onehot = monai.networks.one_hot(interior_maps, num_classes=3)
+
+ return interior_onehot
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/EnsemblePredictor.py b/annolid/segmentation/MEDIAR/core/MEDIAR/EnsemblePredictor.py
new file mode 100644
index 00000000..e65685c7
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/EnsemblePredictor.py
@@ -0,0 +1,105 @@
+import torch
+import os, sys, copy
+import numpy as np
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+from annolid.segmentation.MEDIAR.core.MEDIAR.Predictor import Predictor
+
+__all__ = ["EnsemblePredictor"]
+
+
+class EnsemblePredictor(Predictor):
+ def __init__(
+ self,
+ model,
+ model_aux,
+ device,
+ input_path,
+ output_path,
+ make_submission=False,
+ exp_name=None,
+ algo_params=None,
+ ):
+ super(EnsemblePredictor, self).__init__(
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission,
+ exp_name,
+ algo_params,
+ )
+ self.model_aux = model_aux
+
+ @torch.no_grad()
+ def _inference(self, img_data):
+
+ self.model_aux.to(self.device)
+ self.model_aux.eval()
+
+ img_data = img_data.to(self.device)
+ img_base = img_data
+
+ outputs_base = self._window_inference(img_base)
+ outputs_base = outputs_base.cpu().squeeze()
+
+ outputs_aux = self._window_inference(img_base, aux=True)
+ outputs_aux = outputs_aux.cpu().squeeze()
+ img_base.cpu()
+
+ if not self.use_tta:
+ pred_mask = (outputs_base + outputs_aux) / 2
+ return pred_mask
+
+ else:
+ # HorizontalFlip TTA
+ img_hflip = self.hflip_tta.apply_aug_image(img_data, apply=True)
+
+ outputs_hflip = self._window_inference(img_hflip)
+ outputs_hflip_aux = self._window_inference(img_hflip, aux=True)
+
+ outputs_hflip = self.hflip_tta.apply_deaug_mask(outputs_hflip, apply=True)
+ outputs_hflip_aux = self.hflip_tta.apply_deaug_mask(
+ outputs_hflip_aux, apply=True
+ )
+
+ outputs_hflip = outputs_hflip.cpu().squeeze()
+ outputs_hflip_aux = outputs_hflip_aux.cpu().squeeze()
+ img_hflip = img_hflip.cpu()
+
+ # VertricalFlip TTA
+ img_vflip = self.vflip_tta.apply_aug_image(img_data, apply=True)
+
+ outputs_vflip = self._window_inference(img_vflip)
+ outputs_vflip_aux = self._window_inference(img_vflip, aux=True)
+
+ outputs_vflip = self.vflip_tta.apply_deaug_mask(outputs_vflip, apply=True)
+ outputs_vflip_aux = self.vflip_tta.apply_deaug_mask(
+ outputs_vflip_aux, apply=True
+ )
+
+ outputs_vflip = outputs_vflip.cpu().squeeze()
+ outputs_vflip_aux = outputs_vflip_aux.cpu().squeeze()
+ img_vflip = img_vflip.cpu()
+
+ # Merge Results
+ pred_mask = torch.zeros_like(outputs_base)
+ pred_mask[0] = (outputs_base[0] + outputs_hflip[0] - outputs_vflip[0]) / 3
+ pred_mask[1] = (outputs_base[1] - outputs_hflip[1] + outputs_vflip[1]) / 3
+ pred_mask[2] = (outputs_base[2] + outputs_hflip[2] + outputs_vflip[2]) / 3
+
+ pred_mask_aux = torch.zeros_like(outputs_aux)
+ pred_mask_aux[0] = (
+ outputs_aux[0] + outputs_hflip_aux[0] - outputs_vflip_aux[0]
+ ) / 3
+ pred_mask_aux[1] = (
+ outputs_aux[1] - outputs_hflip_aux[1] + outputs_vflip_aux[1]
+ ) / 3
+ pred_mask_aux[2] = (
+ outputs_aux[2] + outputs_hflip_aux[2] + outputs_vflip_aux[2]
+ ) / 3
+
+ pred_mask = (pred_mask + pred_mask_aux) / 2
+
+ return pred_mask
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/Predictor.py b/annolid/segmentation/MEDIAR/core/MEDIAR/Predictor.py
new file mode 100644
index 00000000..da3f12eb
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/Predictor.py
@@ -0,0 +1,234 @@
+import torch
+import numpy as np
+import os, sys
+from monai.inferers import sliding_window_inference
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+from annolid.segmentation.MEDIAR.core.BasePredictor import BasePredictor
+from annolid.segmentation.MEDIAR.core.MEDIAR.utils import compute_masks
+
+__all__ = ["Predictor"]
+
+
+class Predictor(BasePredictor):
+ def __init__(
+ self,
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission=False,
+ exp_name=None,
+ algo_params=None,
+ ):
+ super(Predictor, self).__init__(
+ model,
+ device,
+ input_path,
+ output_path,
+ make_submission,
+ exp_name,
+ algo_params,
+ )
+ self.hflip_tta = HorizontalFlip()
+ self.vflip_tta = VerticalFlip()
+
+ @torch.no_grad()
+ def _inference(self, img_data):
+ """Conduct model prediction"""
+
+ img_data = img_data.to(self.device)
+ img_base = img_data
+ outputs_base = self._window_inference(img_base)
+ outputs_base = outputs_base.cpu().squeeze()
+ img_base.cpu()
+
+ if not self.use_tta:
+ pred_mask = outputs_base
+ return pred_mask
+
+ else:
+ # HorizontalFlip TTA
+ img_hflip = self.hflip_tta.apply_aug_image(img_data, apply=True)
+ outputs_hflip = self._window_inference(img_hflip)
+ outputs_hflip = self.hflip_tta.apply_deaug_mask(outputs_hflip, apply=True)
+ outputs_hflip = outputs_hflip.cpu().squeeze()
+ img_hflip = img_hflip.cpu()
+
+ # VertricalFlip TTA
+ img_vflip = self.vflip_tta.apply_aug_image(img_data, apply=True)
+ outputs_vflip = self._window_inference(img_vflip)
+ outputs_vflip = self.vflip_tta.apply_deaug_mask(outputs_vflip, apply=True)
+ outputs_vflip = outputs_vflip.cpu().squeeze()
+ img_vflip = img_vflip.cpu()
+
+ # Merge Results
+ pred_mask = torch.zeros_like(outputs_base)
+ pred_mask[0] = (outputs_base[0] + outputs_hflip[0] - outputs_vflip[0]) / 3
+ pred_mask[1] = (outputs_base[1] - outputs_hflip[1] + outputs_vflip[1]) / 3
+ pred_mask[2] = (outputs_base[2] + outputs_hflip[2] + outputs_vflip[2]) / 3
+
+ return pred_mask
+
+ def _window_inference(self, img_data, aux=False):
+ """Inference on RoI-sized window"""
+ outputs = sliding_window_inference(
+ img_data,
+ roi_size=512,
+ sw_batch_size=4,
+ predictor=self.model if not aux else self.model_aux,
+ padding_mode="constant",
+ mode="gaussian",
+ overlap=0.6,
+ )
+
+ return outputs
+
+ def _post_process(self, pred_mask):
+ """Generate cell instance masks."""
+ dP, cellprob = pred_mask[:2], self._sigmoid(pred_mask[-1])
+ H, W = pred_mask.shape[-2], pred_mask.shape[-1]
+
+ if np.prod(H * W) < (5000 * 5000):
+ pred_mask = compute_masks(
+ dP,
+ cellprob,
+ use_gpu=True,
+ flow_threshold=0.4,
+ device=self.device,
+ cellprob_threshold=0.5,
+ )[0]
+
+ else:
+ print("\n[Whole Slide] Grid Prediction starting...")
+ roi_size = 2000
+
+ # Get patch grid by roi_size
+ if H % roi_size != 0:
+ n_H = H // roi_size + 1
+ new_H = roi_size * n_H
+ else:
+ n_H = H // roi_size
+ new_H = H
+
+ if W % roi_size != 0:
+ n_W = W // roi_size + 1
+ new_W = roi_size * n_W
+ else:
+ n_W = W // roi_size
+ new_W = W
+
+ # Allocate values on the grid
+ pred_pad = np.zeros((new_H, new_W), dtype=np.uint32)
+ dP_pad = np.zeros((2, new_H, new_W), dtype=np.float32)
+ cellprob_pad = np.zeros((new_H, new_W), dtype=np.float32)
+
+ dP_pad[:, :H, :W], cellprob_pad[:H, :W] = dP, cellprob
+
+ for i in range(n_H):
+ for j in range(n_W):
+ print("Pred on Grid (%d, %d) processing..." % (i, j))
+ dP_roi = dP_pad[
+ :,
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ]
+ cellprob_roi = cellprob_pad[
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ]
+
+ pred_mask = compute_masks(
+ dP_roi,
+ cellprob_roi,
+ use_gpu=True,
+ flow_threshold=0.4,
+ device=self.device,
+ cellprob_threshold=0.5,
+ )[0]
+
+ pred_pad[
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ] = pred_mask
+
+ pred_mask = pred_pad[:H, :W]
+
+ return pred_mask
+
+ def _sigmoid(self, z):
+ return 1 / (1 + np.exp(-z))
+
+
+"""
+Adapted from the following references:
+[1] https://github.com/qubvel/ttach/blob/master/ttach/transforms.py
+
+"""
+
+
+def hflip(x):
+ """flip batch of images horizontally"""
+ return x.flip(3)
+
+
+def vflip(x):
+ """flip batch of images vertically"""
+ return x.flip(2)
+
+
+class DualTransform:
+ identity_param = None
+
+ def __init__(
+ self, name: str, params,
+ ):
+ self.params = params
+ self.pname = name
+
+ def apply_aug_image(self, image, *args, **params):
+ raise NotImplementedError
+
+ def apply_deaug_mask(self, mask, *args, **params):
+ raise NotImplementedError
+
+
+class HorizontalFlip(DualTransform):
+ """Flip images horizontally (left -> right)"""
+
+ identity_param = False
+
+ def __init__(self):
+ super().__init__("apply", [False, True])
+
+ def apply_aug_image(self, image, apply=False, **kwargs):
+ if apply:
+ image = hflip(image)
+ return image
+
+ def apply_deaug_mask(self, mask, apply=False, **kwargs):
+ if apply:
+ mask = hflip(mask)
+ return mask
+
+
+class VerticalFlip(DualTransform):
+ """Flip images vertically (up -> down)"""
+
+ identity_param = False
+
+ def __init__(self):
+ super().__init__("apply", [False, True])
+
+ def apply_aug_image(self, image, apply=False, **kwargs):
+ if apply:
+ image = vflip(image)
+
+ return image
+
+ def apply_deaug_mask(self, mask, apply=False, **kwargs):
+ if apply:
+ mask = vflip(mask)
+
+ return mask
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/Trainer.py b/annolid/segmentation/MEDIAR/core/MEDIAR/Trainer.py
new file mode 100644
index 00000000..4661f339
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/Trainer.py
@@ -0,0 +1,172 @@
+import torch
+import torch.nn as nn
+import numpy as np
+import os, sys
+from tqdm import tqdm
+from monai.inferers import sliding_window_inference
+
+sys.path.insert(0, os.path.abspath(os.path.join(os.getcwd(), "../../")))
+
+from annolid.segmentation.MEDIAR.core.BaseTrainer import BaseTrainer
+from annolid.segmentation.MEDIAR.core.MEDIAR.utils import *
+
+__all__ = ["Trainer"]
+
+
+class Trainer(BaseTrainer):
+ def __init__(
+ self,
+ model,
+ dataloaders,
+ optimizer,
+ scheduler=None,
+ criterion=None,
+ num_epochs=100,
+ device="cuda:0",
+ no_valid=False,
+ valid_frequency=1,
+ amp=False,
+ algo_params=None,
+ ):
+ super(Trainer, self).__init__(
+ model,
+ dataloaders,
+ optimizer,
+ scheduler,
+ criterion,
+ num_epochs,
+ device,
+ no_valid,
+ valid_frequency,
+ amp,
+ algo_params,
+ )
+
+ self.mse_loss = nn.MSELoss(reduction="mean")
+ self.bce_loss = nn.BCEWithLogitsLoss(reduction="mean")
+
+ def mediar_criterion(self, outputs, labels_onehot_flows):
+ """loss function between true labels and prediction outputs"""
+
+ # Cell Recognition Loss
+ cellprob_loss = self.bce_loss(
+ outputs[:, -1],
+ torch.from_numpy(labels_onehot_flows[:, 1] > 0.5).to(self.device).float(),
+ )
+
+ # Cell Distinction Loss
+ gradient_flows = torch.from_numpy(labels_onehot_flows[:, 2:]).to(self.device)
+ gradflow_loss = 0.5 * self.mse_loss(outputs[:, :2], 5.0 * gradient_flows)
+
+ loss = cellprob_loss + gradflow_loss
+
+ return loss
+
+ def _epoch_phase(self, phase):
+ phase_results = {}
+
+ # Set model mode
+ self.model.train() if phase == "train" else self.model.eval()
+
+ # Epoch process
+ for batch_data in tqdm(self.dataloaders[phase]):
+ images, labels = batch_data["img"], batch_data["label"]
+
+ if self.with_public:
+ # Load batches sequentially from the unlabeled dataloader
+ try:
+ batch_data = next(self.public_iterator)
+ images_pub, labels_pub = batch_data["img"], batch_data["label"]
+
+ except:
+ # Assign memory loader if the cycle ends
+ self.public_iterator = iter(self.public_loader)
+ batch_data = next(self.public_iterator)
+ images_pub, labels_pub = batch_data["img"], batch_data["label"]
+
+ # Concat memory data to the batch
+ images = torch.cat([images, images_pub], dim=0)
+ labels = torch.cat([labels, labels_pub], dim=0)
+
+ images = images.to(self.device)
+ labels = labels.to(self.device)
+
+ self.optimizer.zero_grad()
+
+ # Forward pass
+ with torch.cuda.amp.autocast(enabled=self.amp):
+ with torch.set_grad_enabled(phase == "train"):
+ # Output shape is B x [grad y, grad x, cellprob] x H x W
+ outputs = self._inference(images, phase)
+
+ # Map label masks to graidnet and onehot
+ labels_onehot_flows = labels_to_flows(
+ labels, use_gpu=True, device=self.device
+ )
+ # Calculate loss
+ loss = self.mediar_criterion(outputs, labels_onehot_flows)
+ self.loss_metric.append(loss)
+
+ # Calculate valid statistics
+ if phase != "train":
+ outputs, labels = self._post_process(outputs, labels)
+ f1_score = self._get_f1_metric(outputs, labels)
+ self.f1_metric.append(f1_score)
+
+ # Backward pass
+ if phase == "train":
+ # For the mixed precision training
+ if self.amp:
+ self.scaler.scale(loss).backward()
+ self.scaler.unscale_(self.optimizer)
+ self.scaler.step(self.optimizer)
+ self.scaler.update()
+
+ else:
+ loss.backward()
+ self.optimizer.step()
+
+ # Update metrics
+ phase_results = self._update_results(
+ phase_results, self.loss_metric, "dice_loss", phase
+ )
+ if phase != "train":
+ phase_results = self._update_results(
+ phase_results, self.f1_metric, "f1_score", phase
+ )
+
+ return phase_results
+
+ def _inference(self, images, phase="train"):
+ """inference methods for different phase"""
+
+ if phase != "train":
+ outputs = sliding_window_inference(
+ images,
+ roi_size=512,
+ sw_batch_size=4,
+ predictor=self.model,
+ padding_mode="constant",
+ mode="gaussian",
+ overlap=0.5,
+ )
+ else:
+ outputs = self.model(images)
+
+ return outputs
+
+ def _post_process(self, outputs, labels=None):
+ """Predict cell instances using the gradient tracking"""
+ outputs = outputs.squeeze(0).cpu().numpy()
+ gradflows, cellprob = outputs[:2], self._sigmoid(outputs[-1])
+ outputs = compute_masks(gradflows, cellprob, use_gpu=True, device=self.device)
+ outputs = outputs[0] # (1, C, H, W) -> (C, H, W)
+
+ if labels is not None:
+ labels = labels.squeeze(0).squeeze(0).cpu().numpy()
+
+ return outputs, labels
+
+ def _sigmoid(self, z):
+ """Sigmoid function for numpy arrays"""
+ return 1 / (1 + np.exp(-z))
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/__init__.py b/annolid/segmentation/MEDIAR/core/MEDIAR/__init__.py
new file mode 100644
index 00000000..80cc69b9
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/__init__.py
@@ -0,0 +1,3 @@
+from .Trainer import *
+from .Predictor import *
+from .EnsemblePredictor import *
diff --git a/annolid/segmentation/MEDIAR/core/MEDIAR/utils.py b/annolid/segmentation/MEDIAR/core/MEDIAR/utils.py
new file mode 100644
index 00000000..555ea4e1
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/MEDIAR/utils.py
@@ -0,0 +1,429 @@
+"""
+Copyright © 2022 Howard Hughes Medical Institute,
+Authored by Carsen Stringer and Marius Pachitariu.
+
+Redistribution and use in source and binary forms, with or without
+modification, are permitted provided that the following conditions are met:
+
+1. Redistributions of source code must retain the above copyright notice,
+ this list of conditions and the following disclaimer.
+
+2. Redistributions in binary form must reproduce the above copyright notice,
+ this list of conditions and the following disclaimer in the documentation
+ and/or other materials provided with the distribution.
+
+3. Neither the name of HHMI nor the names of its contributors may be used to
+ endorse or promote products derived from this software without specific
+ prior written permission.
+
+THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
+AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
+IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE
+ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT OWNER OR CONTRIBUTORS BE
+LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR
+CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF
+SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS
+INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN
+CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE)
+ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE
+POSSIBILITY OF SUCH DAMAGE.
+
+--------------------------------------------------------------------------
+MEDIAR Prediction uses CellPose's Gradient Flow Tracking.
+
+This code is adapted from the following codes:
+[1] https://github.com/MouseLand/cellpose/blob/main/cellpose/utils.py
+[2] https://github.com/MouseLand/cellpose/blob/main/cellpose/dynamics.py
+[3] https://github.com/MouseLand/cellpose/blob/main/cellpose/metrics.py
+"""
+
+import torch
+from torch.nn.functional import grid_sample
+import numpy as np
+import fastremap
+
+from skimage import morphology
+from scipy.ndimage import mean, find_objects
+from scipy.ndimage.filters import maximum_filter1d
+
+torch_GPU = torch.device("cuda")
+torch_CPU = torch.device("cpu")
+
+
+def labels_to_flows(labels, use_gpu=False, device=None, redo_flows=False):
+ """
+ Convert labels (list of masks or flows) to flows for training model
+ """
+
+ # Labels b x 1 x h x w
+ labels = labels.cpu().numpy().astype(np.int16)
+ nimg = len(labels)
+
+ if labels[0].ndim < 3:
+ labels = [labels[n][np.newaxis, :, :] for n in range(nimg)]
+
+ # Flows need to be recomputed
+ if labels[0].shape[0] == 1 or labels[0].ndim < 3 or redo_flows:
+ # compute flows; labels are fixed here to be unique, so they need to be passed back
+ # make sure labels are unique!
+ labels = [fastremap.renumber(label, in_place=True)[0] for label in labels]
+ veci = [
+ masks_to_flows(labels[n][0], use_gpu=use_gpu, device=device)
+ for n in range(nimg)
+ ]
+
+ # concatenate labels, distance transform, vector flows, heat (boundary and mask are computed in augmentations)
+ flows = [
+ np.concatenate((labels[n], labels[n] > 0.5, veci[n]), axis=0).astype(
+ np.float32
+ )
+ for n in range(nimg)
+ ]
+
+ return np.array(flows)
+
+
+def compute_masks(
+ dP,
+ cellprob,
+ p=None,
+ niter=200,
+ cellprob_threshold=0.4,
+ flow_threshold=0.4,
+ interp=True,
+ resize=None,
+ use_gpu=False,
+ device=None,
+):
+ """compute masks using dynamics from dP, cellprob, and boundary"""
+
+ cp_mask = cellprob > cellprob_threshold
+ cp_mask = morphology.remove_small_holes(cp_mask, area_threshold=16)
+ cp_mask = morphology.remove_small_objects(cp_mask, min_size=16)
+
+ if np.any(cp_mask): # mask at this point is a cell cluster binary map, not labels
+ # follow flows
+ if p is None:
+ p, inds = follow_flows(
+ dP * cp_mask / 5.0,
+ niter=niter,
+ interp=interp,
+ use_gpu=use_gpu,
+ device=device,
+ )
+ if inds is None:
+ shape = resize if resize is not None else cellprob.shape
+ mask = np.zeros(shape, np.uint16)
+ p = np.zeros((len(shape), *shape), np.uint16)
+ return mask, p
+
+ # calculate masks
+ mask = get_masks(p, iscell=cp_mask)
+
+ # flow thresholding factored out of get_masks
+ shape0 = p.shape[1:]
+ if mask.max() > 0 and flow_threshold is not None and flow_threshold > 0:
+ # make sure labels are unique at output of get_masks
+ mask = remove_bad_flow_masks(
+ mask, dP, threshold=flow_threshold, use_gpu=use_gpu, device=device
+ )
+ else: # nothing to compute, just make it compatible
+ shape = resize if resize is not None else cellprob.shape
+ mask = np.zeros(shape, np.uint16)
+ p = np.zeros((len(shape), *shape), np.uint16)
+
+ return mask, p
+
+
+def _extend_centers_gpu(
+ neighbors, centers, isneighbor, Ly, Lx, n_iter=200, device=torch.device("cuda")
+):
+ if device is not None:
+ device = device
+ nimg = neighbors.shape[0] // 9
+ pt = torch.from_numpy(neighbors).to(device)
+
+ T = torch.zeros((nimg, Ly, Lx), dtype=torch.double, device=device)
+ meds = torch.from_numpy(centers.astype(int)).to(device).long()
+ isneigh = torch.from_numpy(isneighbor).to(device)
+ for i in range(n_iter):
+ T[:, meds[:, 0], meds[:, 1]] += 1
+ Tneigh = T[:, pt[:, :, 0], pt[:, :, 1]]
+ Tneigh *= isneigh
+ T[:, pt[0, :, 0], pt[0, :, 1]] = Tneigh.mean(axis=1)
+ del meds, isneigh, Tneigh
+ T = torch.log(1.0 + T)
+ # gradient positions
+ grads = T[:, pt[[2, 1, 4, 3], :, 0], pt[[2, 1, 4, 3], :, 1]]
+ del pt
+ dy = grads[:, 0] - grads[:, 1]
+ dx = grads[:, 2] - grads[:, 3]
+ del grads
+ mu_torch = np.stack((dy.cpu().squeeze(), dx.cpu().squeeze()), axis=-2)
+ return mu_torch
+
+
+def diameters(masks):
+ _, counts = np.unique(np.int32(masks), return_counts=True)
+ counts = counts[1:]
+ md = np.median(counts ** 0.5)
+ if np.isnan(md):
+ md = 0
+ md /= (np.pi ** 0.5) / 2
+ return md, counts ** 0.5
+
+
+def masks_to_flows_gpu(masks, device=None):
+ if device is None:
+ device = torch.device("cuda")
+
+ Ly0, Lx0 = masks.shape
+ Ly, Lx = Ly0 + 2, Lx0 + 2
+
+ masks_padded = np.zeros((Ly, Lx), np.int64)
+ masks_padded[1:-1, 1:-1] = masks
+
+ # get mask pixel neighbors
+ y, x = np.nonzero(masks_padded)
+ neighborsY = np.stack((y, y - 1, y + 1, y, y, y - 1, y - 1, y + 1, y + 1), axis=0)
+ neighborsX = np.stack((x, x, x, x - 1, x + 1, x - 1, x + 1, x - 1, x + 1), axis=0)
+ neighbors = np.stack((neighborsY, neighborsX), axis=-1)
+
+ # get mask centers
+ slices = find_objects(masks)
+
+ centers = np.zeros((masks.max(), 2), "int")
+ for i, si in enumerate(slices):
+ if si is not None:
+ sr, sc = si
+
+ ly, lx = sr.stop - sr.start + 1, sc.stop - sc.start + 1
+ yi, xi = np.nonzero(masks[sr, sc] == (i + 1))
+ yi = yi.astype(np.int32) + 1 # add padding
+ xi = xi.astype(np.int32) + 1 # add padding
+ ymed = np.median(yi)
+ xmed = np.median(xi)
+ imin = np.argmin((xi - xmed) ** 2 + (yi - ymed) ** 2)
+ xmed = xi[imin]
+ ymed = yi[imin]
+ centers[i, 0] = ymed + sr.start
+ centers[i, 1] = xmed + sc.start
+
+ # get neighbor validator (not all neighbors are in same mask)
+ neighbor_masks = masks_padded[neighbors[:, :, 0], neighbors[:, :, 1]]
+ isneighbor = neighbor_masks == neighbor_masks[0]
+ ext = np.array(
+ [[sr.stop - sr.start + 1, sc.stop - sc.start + 1] for sr, sc in slices]
+ )
+ n_iter = 2 * (ext.sum(axis=1)).max()
+ # run diffusion
+ mu = _extend_centers_gpu(
+ neighbors, centers, isneighbor, Ly, Lx, n_iter=n_iter, device=device
+ )
+
+ # normalize
+ mu /= 1e-20 + (mu ** 2).sum(axis=0) ** 0.5
+
+ # put into original image
+ mu0 = np.zeros((2, Ly0, Lx0))
+ mu0[:, y - 1, x - 1] = mu
+ mu_c = np.zeros_like(mu0)
+ return mu0, mu_c
+
+
+def masks_to_flows(masks, use_gpu=False, device=None):
+ if masks.max() == 0 or (masks != 0).sum() == 1:
+ # dynamics_logger.warning('empty masks!')
+ return np.zeros((2, *masks.shape), "float32")
+
+ if use_gpu:
+ if use_gpu and device is None:
+ device = torch_GPU
+ elif device is None:
+ device = torch_CPU
+ masks_to_flows_device = masks_to_flows_gpu
+
+ if masks.ndim == 3:
+ Lz, Ly, Lx = masks.shape
+ mu = np.zeros((3, Lz, Ly, Lx), np.float32)
+ for z in range(Lz):
+ mu0 = masks_to_flows_device(masks[z], device=device)[0]
+ mu[[1, 2], z] += mu0
+ for y in range(Ly):
+ mu0 = masks_to_flows_device(masks[:, y], device=device)[0]
+ mu[[0, 2], :, y] += mu0
+ for x in range(Lx):
+ mu0 = masks_to_flows_device(masks[:, :, x], device=device)[0]
+ mu[[0, 1], :, :, x] += mu0
+ return mu
+ elif masks.ndim == 2:
+ mu, mu_c = masks_to_flows_device(masks, device=device)
+ return mu
+
+ else:
+ raise ValueError("masks_to_flows only takes 2D or 3D arrays")
+
+
+def steps2D_interp(p, dP, niter, use_gpu=False, device=None):
+ shape = dP.shape[1:]
+ if use_gpu:
+ if device is None:
+ device = torch_GPU
+ shape = (
+ np.array(shape)[[1, 0]].astype("float") - 1
+ ) # Y and X dimensions (dP is 2.Ly.Lx), flipped X-1, Y-1
+ pt = (
+ torch.from_numpy(p[[1, 0]].T).float().to(device).unsqueeze(0).unsqueeze(0)
+ ) # p is n_points by 2, so pt is [1 1 2 n_points]
+ im = (
+ torch.from_numpy(dP[[1, 0]]).float().to(device).unsqueeze(0)
+ ) # covert flow numpy array to tensor on GPU, add dimension
+ # normalize pt between 0 and 1, normalize the flow
+ for k in range(2):
+ im[:, k, :, :] *= 2.0 / shape[k]
+ pt[:, :, :, k] /= shape[k]
+
+ # normalize to between -1 and 1
+ pt = pt * 2 - 1
+
+ # here is where the stepping happens
+ for t in range(niter):
+ # align_corners default is False, just added to suppress warning
+ dPt = grid_sample(im, pt, align_corners=False)
+
+ for k in range(2): # clamp the final pixel locations
+ pt[:, :, :, k] = torch.clamp(
+ pt[:, :, :, k] + dPt[:, k, :, :], -1.0, 1.0
+ )
+
+ # undo the normalization from before, reverse order of operations
+ pt = (pt + 1) * 0.5
+ for k in range(2):
+ pt[:, :, :, k] *= shape[k]
+
+ p = pt[:, :, :, [1, 0]].cpu().numpy().squeeze().T
+ return p
+
+ else:
+ assert print("ho")
+
+
+def follow_flows(dP, mask=None, niter=200, interp=True, use_gpu=True, device=None):
+ shape = np.array(dP.shape[1:]).astype(np.int32)
+ niter = np.uint32(niter)
+
+ p = np.meshgrid(np.arange(shape[0]), np.arange(shape[1]), indexing="ij")
+ p = np.array(p).astype(np.float32)
+
+ inds = np.array(np.nonzero(np.abs(dP[0]) > 1e-3)).astype(np.int32).T
+
+ if inds.ndim < 2 or inds.shape[0] < 5:
+ return p, None
+
+ if not interp:
+ assert print("woo")
+
+ else:
+ p_interp = steps2D_interp(
+ p[:, inds[:, 0], inds[:, 1]], dP, niter, use_gpu=use_gpu, device=device
+ )
+ p[:, inds[:, 0], inds[:, 1]] = p_interp
+
+ return p, inds
+
+
+def flow_error(maski, dP_net, use_gpu=False, device=None):
+ if dP_net.shape[1:] != maski.shape:
+ print("ERROR: net flow is not same size as predicted masks")
+ return
+
+ # flows predicted from estimated masks
+ dP_masks = masks_to_flows(maski, use_gpu=use_gpu, device=device)
+ # difference between predicted flows vs mask flows
+ flow_errors = np.zeros(maski.max())
+ for i in range(dP_masks.shape[0]):
+ flow_errors += mean(
+ (dP_masks[i] - dP_net[i] / 5.0) ** 2,
+ maski,
+ index=np.arange(1, maski.max() + 1),
+ )
+
+ return flow_errors, dP_masks
+
+
+def remove_bad_flow_masks(masks, flows, threshold=0.4, use_gpu=False, device=None):
+ merrors, _ = flow_error(masks, flows, use_gpu, device)
+ badi = 1 + (merrors > threshold).nonzero()[0]
+ masks[np.isin(masks, badi)] = 0
+ return masks
+
+
+def get_masks(p, iscell=None, rpad=20):
+ pflows = []
+ edges = []
+ shape0 = p.shape[1:]
+ dims = len(p)
+
+ for i in range(dims):
+ pflows.append(p[i].flatten().astype("int32"))
+ edges.append(np.arange(-0.5 - rpad, shape0[i] + 0.5 + rpad, 1))
+
+ h, _ = np.histogramdd(tuple(pflows), bins=edges)
+ hmax = h.copy()
+ for i in range(dims):
+ hmax = maximum_filter1d(hmax, 5, axis=i)
+
+ seeds = np.nonzero(np.logical_and(h - hmax > -1e-6, h > 10))
+ Nmax = h[seeds]
+ isort = np.argsort(Nmax)[::-1]
+ for s in seeds:
+ s = s[isort]
+
+ pix = list(np.array(seeds).T)
+
+ shape = h.shape
+ if dims == 3:
+ expand = np.nonzero(np.ones((3, 3, 3)))
+ else:
+ expand = np.nonzero(np.ones((3, 3)))
+ for e in expand:
+ e = np.expand_dims(e, 1)
+
+ for iter in range(5):
+ for k in range(len(pix)):
+ if iter == 0:
+ pix[k] = list(pix[k])
+ newpix = []
+ iin = []
+ for i, e in enumerate(expand):
+ epix = e[:, np.newaxis] + np.expand_dims(pix[k][i], 0) - 1
+ epix = epix.flatten()
+ iin.append(np.logical_and(epix >= 0, epix < shape[i]))
+ newpix.append(epix)
+ iin = np.all(tuple(iin), axis=0)
+ for p in newpix:
+ p = p[iin]
+ newpix = tuple(newpix)
+ igood = h[newpix] > 2
+ for i in range(dims):
+ pix[k][i] = newpix[i][igood]
+ if iter == 4:
+ pix[k] = tuple(pix[k])
+
+ M = np.zeros(h.shape, np.uint32)
+ for k in range(len(pix)):
+ M[pix[k]] = 1 + k
+
+ for i in range(dims):
+ pflows[i] = pflows[i] + rpad
+ M0 = M[tuple(pflows)]
+
+ # remove big masks
+ uniq, counts = fastremap.unique(M0, return_counts=True)
+ big = np.prod(shape0) * 0.9
+ bigc = uniq[counts > big]
+ if len(bigc) > 0 and (len(bigc) > 1 or bigc[0] != 0):
+ M0 = fastremap.mask(M0, bigc)
+ fastremap.renumber(M0, in_place=True) # convenient to guarantee non-skipped labels
+ M0 = np.reshape(M0, shape0)
+ return M0
diff --git a/annolid/segmentation/MEDIAR/core/__init__.py b/annolid/segmentation/MEDIAR/core/__init__.py
new file mode 100644
index 00000000..ff516dce
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/__init__.py
@@ -0,0 +1,2 @@
+from .Baseline import *
+from .MEDIAR import *
diff --git a/annolid/segmentation/MEDIAR/core/utils.py b/annolid/segmentation/MEDIAR/core/utils.py
new file mode 100644
index 00000000..5dc93fbc
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/core/utils.py
@@ -0,0 +1,40 @@
+import torch
+#import wandb
+import pprint
+
+__all__ = ["print_learning_device", "print_with_logging"]
+
+
+def print_learning_device(device):
+ """Get and print the learning device information."""
+ if device == "cpu":
+ device_name = device
+
+ else:
+ if isinstance(device, str):
+ device_idx = int(device[-1])
+ elif isinstance(device, torch._device):
+ device_idx = device.index
+
+ device_name = torch.cuda.get_device_name(device_idx)
+
+ print("")
+ print("=" * 50)
+ print("Train start on device: {}".format(device_name))
+ print("=" * 50)
+
+
+def print_with_logging(results, step):
+ """Print and log on the W&B server.
+
+ Args:
+ results (dict): results dictionary
+ step (int): epoch index
+ """
+ # Print the results dictionary
+ pp = pprint.PrettyPrinter(compact=True)
+ pp.pprint(results)
+ print()
+
+ # Log on the w&b server
+ #wandb.log(results, step=step)
diff --git a/annolid/segmentation/MEDIAR/download_weights.py b/annolid/segmentation/MEDIAR/download_weights.py
new file mode 100644
index 00000000..649d99a1
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/download_weights.py
@@ -0,0 +1,26 @@
+import os
+import gdown
+
+
+def download_weights(output_dir="./weights"):
+ os.makedirs(output_dir, exist_ok=True)
+
+ weights_urls = {
+ "from_phase1": "https://drive.google.com/uc?id=168MtudjTMLoq9YGTyoD2Rjl_d3Gy6c_L",
+ "from_phase2": "https://drive.google.com/uc?id=1JJ2-QKTCk-G7sp5ddkqcifMxgnyOrXjx"
+ }
+
+ for name, url in weights_urls.items():
+ output_path = os.path.join(output_dir, f"{name}.pth")
+ gdown.download(url, output_path, quiet=False)
+
+ return {
+ "model_path1": os.path.join(output_dir, "from_phase1.pth"),
+ "model_path2": os.path.join(output_dir, "from_phase2.pth")
+ }
+
+
+# Example usage:
+weights_paths = download_weights()
+# model_path1 = weights_paths["model_path1"]
+# model_path2 = weights_paths["model_path2"]
diff --git a/annolid/segmentation/MEDIAR/evaluate.py b/annolid/segmentation/MEDIAR/evaluate.py
new file mode 100644
index 00000000..b2a15930
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/evaluate.py
@@ -0,0 +1,71 @@
+import numpy as np
+import pandas as pd
+import tifffile as tif
+import argparse
+import os
+from collections import OrderedDict
+from tqdm import tqdm
+
+from annolid.segmentation.MEDIAR.train_tools.measures import evaluate_f1_score_cellseg
+
+
+def main():
+ ### Directory path arguments ###
+ parser = argparse.ArgumentParser("Compute F1 score for cell segmentation results")
+ parser.add_argument(
+ "--gt_path",
+ type=str,
+ help="path to ground truth; file names end with _label.tiff",
+ required=True,
+ )
+ parser.add_argument(
+ "--pred_path", type=str, help="path to segmentation results", required=True
+ )
+ parser.add_argument("--save_path", default=None, help="path where to save metrics")
+
+ args = parser.parse_args()
+
+ # Get files from the paths
+ gt_path, pred_path = args.gt_path, args.pred_path
+ names = sorted(os.listdir(pred_path))
+
+ names_total = []
+ precisions_total, recalls_total, f1_scores_total = [], [], []
+
+ for name in tqdm(names):
+ assert name.endswith(
+ "_label.tiff"
+ ), "The suffix of label name should be _label.tiff"
+
+ # Load the images
+ gt = tif.imread(os.path.join(gt_path, name))
+ pred = tif.imread(os.path.join(pred_path, name))
+
+ # Evaluate metrics
+ precision, recall, f1_score = evaluate_f1_score_cellseg(gt, pred, threshold=0.5)
+
+ names_total.append(name)
+ precisions_total.append(np.round(precision, 4))
+ recalls_total.append(np.round(recall, 4))
+ f1_scores_total.append(np.round(f1_score, 4))
+
+ # Refine data as dataframe
+ cellseg_metric = OrderedDict()
+ cellseg_metric["Names"] = names_total
+ cellseg_metric["Precision"] = precisions_total
+ cellseg_metric["Recall"] = recalls_total
+ cellseg_metric["F1_Score"] = f1_scores_total
+
+ cellseg_metric = pd.DataFrame(cellseg_metric)
+ print("mean F1 Score:", np.mean(cellseg_metric["F1_Score"]))
+
+ # Save results
+ if args.save_path is not None:
+ os.makedirs(args.save_path, exist_ok=True)
+ cellseg_metric.to_csv(
+ os.path.join(args.save_path, "seg_metric.csv"), index=False
+ )
+
+
+if __name__ == "__main__":
+ main()
diff --git a/annolid/segmentation/MEDIAR/generate_mapping.py b/annolid/segmentation/MEDIAR/generate_mapping.py
new file mode 100644
index 00000000..91c166d2
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/generate_mapping.py
@@ -0,0 +1,121 @@
+import os, glob
+import json
+import argparse
+
+
+def public_paths_labeled(root):
+ """Map paths for public datasets as dictionary list"""
+
+ images_raw = sorted(glob.glob(os.path.join(root, "Public/images/*")))
+ labels_raw = sorted(glob.glob(os.path.join(root, "Public/labels/*")))
+
+ data_dicts = []
+
+ for image_path, label_path in zip(images_raw, labels_raw):
+ name1 = image_path.split("/")[-1].split(".")[0]
+ name2 = label_path.split("/")[-1].split("_label")[0]
+ assert name1 == name2
+
+ data_item = {
+ "img": image_path.split("MEDIAR/")[-1],
+ "label": label_path.split("MEDIAR/")[-1],
+ }
+
+ data_dicts.append(data_item)
+
+ map_dict = {"public": data_dicts}
+
+ return map_dict
+
+
+def official_paths_labeled(root):
+ """Map paths for official labeled datasets as dictionary list"""
+
+ image_path = os.path.join(root, "Official/Training/images/*")
+ label_path = os.path.join(root, "Official/Training/labels/*")
+
+ images_raw = sorted(glob.glob(image_path))
+ labels_raw = sorted(glob.glob(label_path))
+ data_dicts = []
+
+ for image_path, label_path in zip(images_raw, labels_raw):
+ name1 = image_path.split("/")[-1].split(".")[0]
+ name2 = label_path.split("/")[-1].split("_label")[0]
+ assert name1 == name2
+
+ data_item = {
+ "img": image_path.split("MEDIAR/")[-1],
+ "label": label_path.split("MEDIAR/")[-1],
+ }
+
+ data_dicts.append(data_item)
+
+ map_dict = {"official": data_dicts}
+
+ return map_dict
+
+
+def official_paths_tuning(root):
+ """Map paths for official tuning datasets as dictionary list"""
+
+ image_path = os.path.join(root, "Official/Tuning/images/*")
+ images_raw = sorted(glob.glob(image_path))
+
+ data_dicts = []
+
+ for image_path in images_raw:
+ data_item = {"img": image_path.split("MEDIAR/")[-1]}
+ data_dicts.append(data_item)
+
+ map_dict = {"official": data_dicts}
+
+ return map_dict
+
+
+def add_mapping_to_json(json_file, map_dict):
+ """Save mapped dictionary as a json file"""
+
+ if not os.path.exists(json_file):
+ with open(json_file, "w") as file:
+ json.dump({}, file)
+
+ with open(json_file, "r") as file:
+ data = json.load(file)
+
+ for map_key, map_item in map_dict.items():
+ if map_key not in data.keys():
+ data[map_key] = map_item
+ else:
+ print('>>> "{}" already exists in path map keys...'.format(map_key))
+
+ with open(json_file, "w") as file:
+ json.dump(data, file)
+
+
+if __name__ == "__main__":
+ # [!Caution] The paths should be overrided for the local environment!
+ parser = argparse.ArgumentParser(description="Mapping files and paths")
+ parser.add_argument("--root", default=".", type=str)
+ args = parser.parse_args()
+
+ MAP_DIR = "./train_tools/data_utils/"
+
+ print("\n----------- Path Mapping for Labeled Data is Started... -----------\n")
+
+ map_labeled = os.path.join(MAP_DIR, "mapping_labeled.json")
+ map_dict = official_paths_labeled(args.root)
+ add_mapping_to_json(map_labeled, map_dict)
+
+ print("\n----------- Path Mapping for Tuning Data is Started... -----------\n")
+
+ map_labeled = os.path.join(MAP_DIR, "mapping_tuning.json")
+ map_dict = official_paths_tuning(args.root)
+ add_mapping_to_json(map_labeled, map_dict)
+
+ print("\n----------- Path Mapping for Public Data is Started... -----------\n")
+
+ map_public = os.path.join(MAP_DIR, "mapping_public.json")
+ map_dict = public_paths_labeled(args.root)
+ add_mapping_to_json(map_public, map_dict)
+
+ print("\n-------------- Path Mapping is Ended !!! ---------------------------\n")
diff --git a/annolid/segmentation/MEDIAR/image/examples/img2.tif b/annolid/segmentation/MEDIAR/image/examples/img2.tif
new file mode 100644
index 00000000..07780c71
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/examples/img2.tif differ
diff --git a/annolid/segmentation/MEDIAR/image/failure_cases.png b/annolid/segmentation/MEDIAR/image/failure_cases.png
new file mode 100644
index 00000000..fd9ab055
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/failure_cases.png differ
diff --git a/annolid/segmentation/MEDIAR/image/mediar_framework.png b/annolid/segmentation/MEDIAR/image/mediar_framework.png
new file mode 100644
index 00000000..8deb7a22
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/mediar_framework.png differ
diff --git a/annolid/segmentation/MEDIAR/image/mediar_model.PNG b/annolid/segmentation/MEDIAR/image/mediar_model.PNG
new file mode 100644
index 00000000..c842f9da
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/mediar_model.PNG differ
diff --git a/annolid/segmentation/MEDIAR/image/mediar_results.png b/annolid/segmentation/MEDIAR/image/mediar_results.png
new file mode 100644
index 00000000..8c05cf39
Binary files /dev/null and b/annolid/segmentation/MEDIAR/image/mediar_results.png differ
diff --git a/annolid/segmentation/MEDIAR/predict.py b/annolid/segmentation/MEDIAR/predict.py
new file mode 100644
index 00000000..02f86385
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/predict.py
@@ -0,0 +1,70 @@
+import torch
+import argparse, pprint
+
+from annolid.segmentation.MEDIAR.train_tools import *
+from annolid.segmentation.MEDIAR.SetupDict import MODELS, PREDICTOR
+
+# Set torch base print precision
+torch.set_printoptions(6)
+
+
+def main(args):
+ """Execute prediction and save the results"""
+
+ model_args = args.pred_setups.model
+ model = MODELS[model_args.name](**model_args.params)
+
+ if "ensemble" in args.pred_setups.name:
+ weights = torch.load(args.pred_setups.model_path1, map_location="cpu")
+ model.load_state_dict(weights, strict=False)
+
+ model_aux = MODELS[model_args.name](**model_args.params)
+ weights_aux = torch.load(args.pred_setups.model_path2, map_location="cpu")
+ model_aux.load_state_dict(weights_aux, strict=False)
+
+ predictor = PREDICTOR[args.pred_setups.name](
+ model,
+ model_aux,
+ args.pred_setups.device,
+ args.pred_setups.input_path,
+ args.pred_setups.output_path,
+ args.pred_setups.make_submission,
+ args.pred_setups.exp_name,
+ args.pred_setups.algo_params,
+ )
+
+ else:
+ weights = torch.load(args.pred_setups.model_path, map_location="cpu")
+ model.load_state_dict(weights, strict=False)
+
+ predictor = PREDICTOR[args.pred_setups.name](
+ model,
+ args.pred_setups.device,
+ args.pred_setups.input_path,
+ args.pred_setups.output_path,
+ args.pred_setups.make_submission,
+ args.pred_setups.exp_name,
+ args.pred_setups.algo_params,
+ )
+
+ _ = predictor.conduct_prediction()
+
+
+# Parser arguments for terminal execution
+parser = argparse.ArgumentParser(description="Config file processing")
+parser.add_argument(
+ "--config_path", default="./config/step3_prediction/base_prediction.json", type=str
+)
+args = parser.parse_args()
+
+#######################################################################################
+
+if __name__ == "__main__":
+ # Load configuration from .json file
+ opt = ConfLoader(args.config_path).opt
+
+ # Print configuration dictionary pretty
+ pprint_config(opt)
+
+ # Run experiment
+ main(opt)
diff --git a/annolid/segmentation/MEDIAR/predict_ensemble.py b/annolid/segmentation/MEDIAR/predict_ensemble.py
new file mode 100644
index 00000000..5edf47d6
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/predict_ensemble.py
@@ -0,0 +1,196 @@
+import os
+import torch
+import time
+from datetime import datetime
+import numpy as np
+from annolid.segmentation.MEDIAR.train_tools import *
+from annolid.segmentation.MEDIAR.train_tools.models import MEDIARFormer
+from annolid.utils.weights import WeightDownloader
+from annolid.segmentation.MEDIAR.core.MEDIAR import EnsemblePredictor
+from annolid.gui.shape import MaskShape
+from annolid.annotation.keypoints import save_labels
+from annolid.segmentation.MEDIAR.train_tools.data_utils.transforms import get_pred_transforms
+from annolid.utils.logger import logger
+
+
+class MEDIARPredictor(EnsemblePredictor):
+ """
+ Class for conducting cell detection using MEDIAR model.
+ https://github.com/Lee-Gihun/MEDIAR
+ @article{lee2022mediar,
+ title={Mediar: Harmony of data-centric and model-centric for multi-modality microscopy},
+ author={Lee, Gihun and Kim, SangMook and Kim, Joonkee and Yun, Se-Young},
+ journal={arXiv preprint arXiv:2212.03465},
+ year={2022}
+ }
+
+ Args:
+ weights_dir (str): Directory to store downloaded weights.
+ input_path (str): Directory containing input images.
+ output_path (str): Directory to store output predictions.
+ """
+
+ def __init__(self,
+ input_path="./data/images",
+ output_path="./output",
+ weights_dir="None",
+ ):
+ self.weights_dir = weights_dir
+ self.input_path = input_path
+ self.output_path = output_path
+ self.model1 = None
+ self.model2 = None
+ self.device = 'cuda' if torch.cuda.is_available() else 'cpu'
+ self.download_weights()
+ self.load_models()
+ super(MEDIARPredictor, self).__init__(self.model1, self.model2,
+ self.device, self.input_path,
+ self.output_path, algo_params={"use_tta": True})
+
+ def _setups(self):
+ self.pred_transforms = get_pred_transforms()
+ os.makedirs(self.output_path, exist_ok=True)
+
+ now = datetime.now()
+ dt_string = now.strftime("%m%d_%H%M")
+ self.exp_name = (
+ self.exp_name + dt_string if self.exp_name is not None else dt_string
+ )
+
+ self.img_names = [img_file for img_file in sorted(
+ os.listdir(self.input_path)) if '.json' not in img_file]
+ logger.info(f"Working on the images: {self.img_names}")
+
+ def download_weights(self, weights_dir=None):
+ """
+ Download pretrained weights for MEDIAR models.
+ """
+ if weights_dir is None:
+ self.weights_dir = os.path.join(
+ os.path.dirname(__file__), "weights")
+ downloader = WeightDownloader(self.weights_dir)
+
+ # Define weight URLs, expected checksums, and file names
+ weight_urls = [
+ "https://drive.google.com/uc?id=168MtudjTMLoq9YGTyoD2Rjl_d3Gy6c_L",
+ "https://drive.google.com/uc?id=1JJ2-QKTCk-G7sp5ddkqcifMxgnyOrXjx"
+ ]
+ expected_checksums = [
+ "e0ccb052828a9f05e21b2143939583c5",
+ "a595336926767afdf1ffb1baffd5ab7f"
+ ]
+ weight_file_names = ["from_phase1.pth", "from_phase2.pth"]
+
+ # Download weights for each URL
+ for url, checksum, file_name in zip(weight_urls, expected_checksums, weight_file_names):
+ downloader.download_weights(url, checksum, file_name)
+
+ def load_models(self):
+ """
+ Load pretrained MEDIAR models.
+ """
+ model_args = {
+ "classes": 3,
+ "decoder_channels": [1024, 512, 256, 128, 64],
+ "decoder_pab_channels": 256,
+ "encoder_name": 'mit_b5',
+ "in_channels": 3
+ }
+ self.model1 = MEDIARFormer(**model_args)
+ self.model1.load_state_dict(torch.load(f"{self.weights_dir}/from_phase1.pth",
+ map_location="cpu"), strict=False)
+
+ self.model2 = MEDIARFormer(**model_args)
+ self.model2.load_state_dict(torch.load(f"{self.weights_dir}/from_phase2.pth",
+ map_location="cpu"), strict=False)
+
+ @torch.no_grad()
+ def conduct_prediction(self):
+ self.model.to(self.device)
+ self.model.eval()
+ total_time = 0
+ total_times = []
+
+ for img_name in self.img_names:
+ img_data = self._get_img_data(img_name)
+ img_data = img_data.to(self.device)
+
+ start = time.time()
+
+ pred_mask = self._inference(img_data)
+ pred_mask = self._post_process(pred_mask.squeeze(0).cpu().numpy())
+
+ self.write_pred_mask(
+ pred_mask, self.output_path, img_name, self.make_submission
+ )
+ shape_list = self.save_prediction(
+ pred_mask, image_name=img_name)
+ end = time.time()
+
+ time_cost = end - start
+ total_times.append(time_cost)
+ total_time += time_cost
+ logger.info(
+ f"Prediction finished: {img_name}; img size = {img_data.shape}; costing: {time_cost:.2f}s"
+ )
+
+ logger.info(f"\n Total Time Cost: {total_time:.2f}s")
+
+ return shape_list
+
+ def _save_annotation(self, filename, mask_dict,
+ frame_shape,
+ img_ext='.png'):
+ if len(frame_shape) == 3:
+ height, width, _ = frame_shape
+ else:
+ height, width = frame_shape
+ label_list = []
+ for label_id, mask in mask_dict.items():
+ label = str(label_id)
+ current_shape = MaskShape(label=label,
+ flags={},
+ description='cell segmentation')
+ current_shape.mask = mask
+ _shapes = current_shape.toPolygons(
+ epsilon=2.0)
+ if len(_shapes) < 0:
+ continue
+ current_shape = _shapes[0]
+ points = [[point.x(), point.y()] for point in current_shape.points]
+ current_shape.points = points
+ label_list.append(current_shape)
+ img_abs_path = filename.replace('.json', img_ext)
+ save_labels(filename=filename,
+ imagePath=img_abs_path,
+ label_list=label_list,
+ height=height,
+ width=width,
+ save_image_to_json=False)
+ return label_list
+
+ def save_prediction(self, pred_mask, image_name="img1"):
+ """
+ Save prediction for a specific image.
+
+ Args:
+ image_name (str): Name of the input image file.
+ """
+ img_filename = os.path.join(self.input_path, image_name)
+ _, ext = os.path.splitext(img_filename)
+ # Replace the extension with ".json"
+ json_filename = img_filename.replace(ext, '.json')
+ json_filename = os.path.abspath(json_filename)
+ mask_dict = {f'cell_{label_id}': (pred_mask == label_id)
+ for label_id in np.unique(pred_mask)[1:]}
+ shape_list = self._save_annotation(json_filename, mask_dict,
+ pred_mask.shape, img_ext=ext)
+
+ cell_count = len(np.unique(pred_mask)) - 1 # exclude the background
+ logger.info(f"\n{cell_count} Cells detected!")
+ return shape_list
+
+
+if __name__ == "__main__":
+ mediar_predictor = MEDIARPredictor()
+ shape_list = mediar_predictor.conduct_prediction()
diff --git a/annolid/segmentation/MEDIAR/requirements.txt b/annolid/segmentation/MEDIAR/requirements.txt
new file mode 100644
index 00000000..7464efd9
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/requirements.txt
@@ -0,0 +1,14 @@
+fastremap==1.14.1
+monai==1.3.0
+numba==0.57.1
+numpy==1.24.3
+pandas==2.0.3
+pytz==2023.3.post1
+scipy==1.12.0
+segmentation_models_pytorch==0.3.3
+tifffile==2023.4.12
+torch==2.1.2
+tqdm==4.65.0
+wandb==0.16.2
+scikit-image
+matplotlib
diff --git a/annolid/segmentation/MEDIAR/train_tools/__init__.py b/annolid/segmentation/MEDIAR/train_tools/__init__.py
new file mode 100644
index 00000000..7614b006
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/__init__.py
@@ -0,0 +1,3 @@
+from .data_utils import *
+from .measures import *
+from .utils import *
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/__init__.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/__init__.py
new file mode 100644
index 00000000..e387c35e
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/__init__.py
@@ -0,0 +1 @@
+from .datasetter import *
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/CellAware.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/CellAware.py
new file mode 100644
index 00000000..24be12f6
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/CellAware.py
@@ -0,0 +1,88 @@
+import numpy as np
+import copy
+
+from monai.transforms import RandScaleIntensity, Compose
+from monai.transforms.compose import MapTransform
+from skimage.segmentation import find_boundaries
+
+
+__all__ = ["BoundaryExclusion", "IntensityDiversification"]
+
+
+class BoundaryExclusion(MapTransform):
+ """Map the cell boundary pixel labels to the background class (0)."""
+
+ def __init__(self, keys=["label"], allow_missing_keys=False):
+ super(BoundaryExclusion, self).__init__(keys, allow_missing_keys)
+
+ def __call__(self, data):
+ # Find and Exclude Boundary
+ label_original = data["label"]
+ label = copy.deepcopy(label_original)
+ boundary = find_boundaries(label, connectivity=1, mode="thick")
+ label[boundary] = 0
+
+ # Do not exclude if the cell is too small (< 14x14).
+ new_label = copy.deepcopy(label_original)
+ new_label[label == 0] = 0
+
+ cell_idx, cell_counts = np.unique(label_original, return_counts=True)
+
+ for k in range(len(cell_counts)):
+ if cell_counts[k] < 196:
+ new_label[label_original == cell_idx[k]] = cell_idx[k]
+
+ # Do not exclude if the pixels are at the image boundaries.
+ _, W, H = label_original.shape
+ bd = np.zeros_like(label_original, dtype=label.dtype)
+ bd[:, 2 : W - 2, 2 : H - 2] = 1
+ new_label += label_original * bd
+
+ # Assign the transformed label
+ data["label"] = new_label
+
+ return data
+
+
+class IntensityDiversification(MapTransform):
+ """Randomly rescale the intensity of cell pixels."""
+
+ def __init__(
+ self,
+ keys=["img"],
+ change_cell_ratio=0.4,
+ scale_factors=[0, 0.7],
+ allow_missing_keys=False,
+ ):
+ super(IntensityDiversification, self).__init__(keys, allow_missing_keys)
+
+ self.change_cell_ratio = change_cell_ratio
+ self.randscale_intensity = Compose(
+ [RandScaleIntensity(prob=1.0, factors=scale_factors)]
+ )
+
+ def __call__(self, data):
+ # Select cells to be transformed
+ cell_count = int(data["label"].max())
+ change_cell_count = int(cell_count * self.change_cell_ratio)
+ change_cell = np.random.choice(cell_count, change_cell_count, replace=False)
+
+ mask = copy.deepcopy(data["label"])
+
+ for i in range(cell_count):
+ cell_id = i + 1
+
+ if cell_id not in change_cell:
+ mask[mask == cell_id] = 0
+
+ mask[mask > 0] = 1
+
+ # Conduct intensity transformation for the selected cells
+ img_original = copy.deepcopy((1 - mask) * data["img"])
+ img_transformed = copy.deepcopy(mask * data["img"])
+ img_transformed = self.randscale_intensity(img_transformed)
+
+ # Assign the transformed image
+ data["img"] = img_original + img_transformed
+
+ return data
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/LoadImage.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/LoadImage.py
new file mode 100644
index 00000000..aad77b13
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/LoadImage.py
@@ -0,0 +1,161 @@
+import numpy as np
+import tifffile as tif
+import skimage.io as io
+from typing import Optional, Sequence, Union
+from monai.config import DtypeLike, PathLike, KeysCollection
+from monai.utils import ensure_tuple
+from monai.data.utils import is_supported_format, optional_import, ensure_tuple_rep
+from monai.data.image_reader import ImageReader, NumpyReader
+from monai.transforms import LoadImage, LoadImaged
+from monai.utils.enums import PostFix
+
+DEFAULT_POST_FIX = PostFix.meta()
+itk, has_itk = optional_import("itk", allow_namespace_pkg=True)
+
+__all__ = [
+ "CustomLoadImaged",
+ "CustomLoadImageD",
+ "CustomLoadImageDict",
+ "CustomLoadImage",
+]
+
+
+class CustomLoadImage(LoadImage):
+ """
+ Load image file or files from provided path based on reader.
+ If reader is not specified, this class automatically chooses readers
+ based on the supported suffixes and in the following order:
+
+ - User-specified reader at runtime when calling this loader.
+ - User-specified reader in the constructor of `LoadImage`.
+ - Readers from the last to the first in the registered list.
+ - Current default readers: (nii, nii.gz -> NibabelReader), (png, jpg, bmp -> PILReader),
+ (npz, npy -> NumpyReader), (nrrd -> NrrdReader), (DICOM file -> ITKReader).
+
+ [!Caution] This overriding replaces the original ITK with Custom UnifiedITKReader.
+ """
+
+ def __init__(
+ self,
+ reader=None,
+ image_only: bool = False,
+ dtype: DtypeLike = np.float32,
+ ensure_channel_first: bool = False,
+ *args,
+ **kwargs,
+ ) -> None:
+ super(CustomLoadImage, self).__init__(
+ reader, image_only, dtype, ensure_channel_first, *args, **kwargs
+ )
+
+ # Adding TIFFReader. Although ITK Reader supports ".tiff" files, sometimes fails to load images.
+ self.readers = []
+ self.register(UnifiedITKReader(*args, **kwargs))
+
+
+class CustomLoadImaged(LoadImaged):
+ """
+ Dictionary-based wrapper of `CustomLoadImage`.
+ """
+
+ def __init__(
+ self,
+ keys: KeysCollection,
+ reader: Optional[Union[ImageReader, str]] = None,
+ dtype: DtypeLike = np.float32,
+ meta_keys: Optional[KeysCollection] = None,
+ meta_key_postfix: str = DEFAULT_POST_FIX,
+ overwriting: bool = False,
+ image_only: bool = False,
+ ensure_channel_first: bool = False,
+ simple_keys=False,
+ allow_missing_keys: bool = False,
+ *args,
+ **kwargs,
+ ) -> None:
+ super(CustomLoadImaged, self).__init__(
+ keys,
+ reader,
+ dtype,
+ meta_keys,
+ meta_key_postfix,
+ overwriting,
+ image_only,
+ ensure_channel_first,
+ simple_keys,
+ allow_missing_keys,
+ *args,
+ **kwargs,
+ )
+
+ # Assign CustomLoader
+ self._loader = CustomLoadImage(
+ reader, image_only, dtype, ensure_channel_first, *args, **kwargs
+ )
+ if not isinstance(meta_key_postfix, str):
+ raise TypeError(
+ f"meta_key_postfix must be a str but is {type(meta_key_postfix).__name__}."
+ )
+ self.meta_keys = (
+ ensure_tuple_rep(None, len(self.keys))
+ if meta_keys is None
+ else ensure_tuple(meta_keys)
+ )
+ if len(self.keys) != len(self.meta_keys):
+ raise ValueError("meta_keys should have the same length as keys.")
+ self.meta_key_postfix = ensure_tuple_rep(meta_key_postfix, len(self.keys))
+ self.overwriting = overwriting
+
+
+class UnifiedITKReader(NumpyReader):
+ """
+ Unified Reader to read ".tif" and ".tiff files".
+ As the tifffile reads the images as numpy arrays, it inherits from the NumpyReader.
+ """
+
+ def __init__(
+ self, channel_dim: Optional[int] = None, **kwargs,
+ ):
+ super(UnifiedITKReader, self).__init__(channel_dim=channel_dim, **kwargs)
+ self.kwargs = kwargs
+ self.channel_dim = channel_dim
+
+ def verify_suffix(self, filename: Union[Sequence[PathLike], PathLike]) -> bool:
+ """Verify whether the file format is supported by TIFF Reader."""
+
+ suffixes: Sequence[str] = ["tif", "tiff", "png", "jpg", "bmp", "jpeg",]
+ return has_itk or is_supported_format(filename, suffixes)
+
+ def read(self, data: Union[Sequence[PathLike], PathLike], **kwargs):
+ """Read Images from the file."""
+ img_ = []
+
+ filenames: Sequence[PathLike] = ensure_tuple(data)
+ kwargs_ = self.kwargs.copy()
+ kwargs_.update(kwargs)
+
+ for name in filenames:
+ name = f"{name}"
+
+ if name.endswith(".tif") or name.endswith(".tiff"):
+ _obj = tif.imread(name)
+ else:
+ try:
+ _obj = itk.imread(name, **kwargs_)
+ _obj = itk.array_view_from_image(_obj, keep_axes=False)
+ except:
+ _obj = io.imread(name)
+
+ if len(_obj.shape) == 2:
+ _obj = np.repeat(np.expand_dims(_obj, axis=-1), 3, axis=-1)
+ elif len(_obj.shape) == 3 and _obj.shape[-1] > 3:
+ _obj = _obj[:, :, :3]
+ else:
+ pass
+
+ img_.append(_obj)
+
+ return img_ if len(filenames) > 1 else img_[0]
+
+
+CustomLoadImageD = CustomLoadImageDict = CustomLoadImaged
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/NormalizeImage.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/NormalizeImage.py
new file mode 100644
index 00000000..08a21509
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/NormalizeImage.py
@@ -0,0 +1,77 @@
+import numpy as np
+from skimage import exposure
+from monai.config import KeysCollection
+
+from monai.transforms.transform import Transform
+from monai.transforms.compose import MapTransform
+
+from typing import Dict, Hashable, Mapping
+
+
+__all__ = [
+ "CustomNormalizeImage",
+ "CustomNormalizeImageD",
+ "CustomNormalizeImageDict",
+ "CustomNormalizeImaged",
+]
+
+
+class CustomNormalizeImage(Transform):
+ """Normalize the image."""
+
+ def __init__(self, percentiles=[0, 99.5], channel_wise=False):
+ self.lower, self.upper = percentiles
+ self.channel_wise = channel_wise
+
+ def _normalize(self, img) -> np.ndarray:
+ non_zero_vals = img[np.nonzero(img)]
+ percentiles = np.percentile(non_zero_vals, [self.lower, self.upper])
+ img_norm = exposure.rescale_intensity(
+ img, in_range=(percentiles[0], percentiles[1]), out_range="uint8"
+ )
+
+ return img_norm.astype(np.uint8)
+
+ def __call__(self, img: np.ndarray) -> np.ndarray:
+ if self.channel_wise:
+ pre_img_data = np.zeros(img.shape, dtype=np.uint8)
+ for i in range(img.shape[-1]):
+ img_channel_i = img[:, :, i]
+
+ if len(img_channel_i[np.nonzero(img_channel_i)]) > 0:
+ pre_img_data[:, :, i] = self._normalize(img_channel_i)
+
+ img = pre_img_data
+
+ else:
+ img = self._normalize(img)
+
+ return img
+
+
+class CustomNormalizeImaged(MapTransform):
+ """Dictionary-based wrapper of NormalizeImage"""
+
+ def __init__(
+ self,
+ keys: KeysCollection,
+ percentiles=[1, 99],
+ channel_wise: bool = False,
+ allow_missing_keys: bool = False,
+ ):
+ super(CustomNormalizeImageD, self).__init__(keys, allow_missing_keys)
+ self.normalizer = CustomNormalizeImage(percentiles, channel_wise)
+
+ def __call__(
+ self, data: Mapping[Hashable, np.ndarray]
+ ) -> Dict[Hashable, np.ndarray]:
+
+ d = dict(data)
+
+ for key in self.keys:
+ d[key] = self.normalizer(d[key])
+
+ return d
+
+
+CustomNormalizeImageD = CustomNormalizeImageDict = CustomNormalizeImaged
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/__init__.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/__init__.py
new file mode 100644
index 00000000..33fc9ebe
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/__init__.py
@@ -0,0 +1,3 @@
+from .LoadImage import *
+from .NormalizeImage import *
+from .CellAware import *
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/modalities.pkl b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/modalities.pkl
new file mode 100644
index 00000000..4f867966
Binary files /dev/null and b/annolid/segmentation/MEDIAR/train_tools/data_utils/custom/modalities.pkl differ
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/datasetter.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/datasetter.py
new file mode 100644
index 00000000..6a6b3317
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/datasetter.py
@@ -0,0 +1,208 @@
+from torch.utils.data import DataLoader
+from monai.data import Dataset
+import pickle
+
+from .transforms import (
+ train_transforms,
+ public_transforms,
+ valid_transforms,
+ tuning_transforms,
+ unlabeled_transforms,
+)
+from .utils import split_train_valid, path_decoder
+
+DATA_LABEL_DICT_PICKLE_FILE = "./train_tools/data_utils/custom/modalities.pkl"
+
+__all__ = [
+ "get_dataloaders_labeled",
+ "get_dataloaders_public",
+ "get_dataloaders_unlabeled",
+]
+
+
+def get_dataloaders_labeled(
+ root,
+ mapping_file,
+ mapping_file_tuning,
+ join_mapping_file=None,
+ valid_portion=0.0,
+ batch_size=8,
+ amplified=False,
+ relabel=False,
+):
+ """Set DataLoaders for labeled datasets.
+
+ Args:
+ root (str): root directory
+ mapping_file (str): json file for mapping dataset
+ valid_portion (float, optional): portion of valid datasets. Defaults to 0.1.
+ batch_size (int, optional): batch size. Defaults to 8.
+ shuffle (bool, optional): shuffles dataloader. Defaults to True.
+ num_workers (int, optional): number of workers for each datalaoder. Defaults to 5.
+
+ Returns:
+ dict: dictionary of data loaders.
+ """
+
+ # Get list of data dictionaries from decoded paths
+ data_dicts = path_decoder(root, mapping_file)
+ tuning_dicts = path_decoder(root, mapping_file_tuning, no_label=True)
+
+ if amplified:
+ with open(DATA_LABEL_DICT_PICKLE_FILE, "rb") as f:
+ data_label_dict = pickle.load(f)
+
+ data_point_dict = {}
+
+ for label, data_lst in data_label_dict.items():
+ data_point_dict[label] = []
+
+ for d_idx in data_lst:
+ try:
+ data_point_dict[label].append(data_dicts[d_idx])
+ except:
+ print(label, d_idx)
+
+ data_dicts = []
+
+ for label, data_points in data_point_dict.items():
+ len_data_points = len(data_points)
+
+ if len_data_points >= 50:
+ data_dicts += data_points
+ else:
+ for i in range(50):
+ data_dicts.append(data_points[i % len_data_points])
+
+ data_transforms = train_transforms
+
+ if join_mapping_file is not None:
+ data_dicts += path_decoder(root, join_mapping_file)
+ data_transforms = public_transforms
+
+ if relabel:
+ for elem in data_dicts:
+ cell_idx = int(elem["label"].split("_label.tiff")[0].split("_")[-1])
+ if cell_idx in range(340, 499):
+ new_label = elem["label"].replace(
+ "/data/CellSeg/Official/Train_Labeled/labels/",
+ "/CellSeg/pretrained_train_ext/",
+ )
+ elem["label"] = new_label
+
+ # Split datasets as Train/Valid
+ train_dicts, valid_dicts = split_train_valid(
+ data_dicts, valid_portion=valid_portion
+ )
+
+ # Obtain datasets with transforms
+ trainset = Dataset(train_dicts, transform=data_transforms)
+ validset = Dataset(valid_dicts, transform=valid_transforms)
+ tuningset = Dataset(tuning_dicts, transform=tuning_transforms)
+
+ # Set dataloader for Trainset
+ train_loader = DataLoader(
+ trainset, batch_size=batch_size, shuffle=True, num_workers=5
+ )
+
+ # Set dataloader for Validset (Batch size is fixed as 1)
+ valid_loader = DataLoader(validset, batch_size=1, shuffle=False,)
+
+ # Set dataloader for Tuningset (Batch size is fixed as 1)
+ tuning_loader = DataLoader(tuningset, batch_size=1, shuffle=False)
+
+ # Form dataloaders as dictionary
+ dataloaders = {
+ "train": train_loader,
+ "valid": valid_loader,
+ "tuning": tuning_loader,
+ }
+
+ return dataloaders
+
+
+def get_dataloaders_public(
+ root, mapping_file, valid_portion=0.0, batch_size=8,
+):
+ """Set DataLoaders for labeled datasets.
+
+ Args:
+ root (str): root directory
+ mapping_file (str): json file for mapping dataset
+ valid_portion (float, optional): portion of valid datasets. Defaults to 0.1.
+ batch_size (int, optional): batch size. Defaults to 8.
+ shuffle (bool, optional): shuffles dataloader. Defaults to True.
+
+ Returns:
+ dict: dictionary of data loaders.
+ """
+
+ # Get list of data dictionaries from decoded paths
+ data_dicts = path_decoder(root, mapping_file)
+
+ # Split datasets as Train/Valid
+ train_dicts, _ = split_train_valid(data_dicts, valid_portion=valid_portion)
+
+ trainset = Dataset(train_dicts, transform=public_transforms)
+ # Set dataloader for Trainset
+ train_loader = DataLoader(
+ trainset, batch_size=batch_size, shuffle=True, num_workers=5
+ )
+
+ # Form dataloaders as dictionary
+ dataloaders = {
+ "public": train_loader,
+ }
+
+ return dataloaders
+
+
+def get_dataloaders_unlabeled(
+ root, mapping_file, batch_size=8, shuffle=True, num_workers=5,
+):
+ """Set dataloaders for unlabeled dataset."""
+ # Get list of data dictionaries from decoded paths
+ unlabeled_dicts = path_decoder(root, mapping_file, no_label=True, unlabeled=True)
+
+ # Obtain datasets with transforms
+ unlabeled_dicts, _ = split_train_valid(unlabeled_dicts, valid_portion=0)
+ unlabeled_set = Dataset(unlabeled_dicts, transform=unlabeled_transforms)
+
+ # Set dataloader for Unlabeled dataset
+ unlabeled_loader = DataLoader(
+ unlabeled_set, batch_size=batch_size, shuffle=shuffle, num_workers=num_workers
+ )
+
+ dataloaders = {
+ "unlabeled": unlabeled_loader,
+ }
+
+ return dataloaders
+
+
+def get_dataloaders_unlabeled_psuedo(
+ root, mapping_file, batch_size=8, shuffle=True, num_workers=5,
+):
+
+ # Get list of data dictionaries from decoded paths
+ unlabeled_psuedo_dicts = path_decoder(
+ root, mapping_file, no_label=False, unlabeled=True
+ )
+
+ # Obtain datasets with transforms
+ unlabeled_psuedo_dicts, _ = split_train_valid(
+ unlabeled_psuedo_dicts, valid_portion=0
+ )
+ unlabeled_psuedo_set = Dataset(unlabeled_psuedo_dicts, transform=train_transforms)
+
+ # Set dataloader for Unlabeled dataset
+ unlabeled_psuedo_loader = DataLoader(
+ unlabeled_psuedo_set,
+ batch_size=batch_size,
+ shuffle=shuffle,
+ num_workers=num_workers,
+ )
+
+ dataloaders = {"unlabeled": unlabeled_psuedo_loader}
+
+ return dataloaders
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/transforms.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/transforms.py
new file mode 100644
index 00000000..00560027
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/transforms.py
@@ -0,0 +1,148 @@
+from .custom import *
+
+from monai.transforms import *
+
+__all__ = [
+ "train_transforms",
+ "public_transforms",
+ "valid_transforms",
+ "tuning_transforms",
+ "unlabeled_transforms",
+]
+
+train_transforms = Compose(
+ [
+ # >>> Load and refine data --- img: (H, W, 3); label: (H, W)
+ CustomLoadImaged(keys=["img", "label"], image_only=True),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img", "label"], channel_dim=-1),
+ RemoveRepeatedChanneld(keys=["label"], repeats=3), # label: (H, W)
+ ScaleIntensityd(keys=["img"], allow_missing_keys=True), # Do not scale label
+ # >>> Spatial transforms
+ RandZoomd(
+ keys=["img", "label"],
+ prob=0.5,
+ min_zoom=0.25,
+ max_zoom=1.5,
+ mode=["area", "nearest"],
+ keep_size=False,
+ ),
+ SpatialPadd(keys=["img", "label"], spatial_size=512),
+ RandSpatialCropd(keys=["img", "label"], roi_size=512, random_size=False),
+ RandAxisFlipd(keys=["img", "label"], prob=0.5),
+ RandRotate90d(keys=["img", "label"], prob=0.5, spatial_axes=[0, 1]),
+ IntensityDiversification(keys=["img", "label"], allow_missing_keys=True),
+ # # >>> Intensity transforms
+ RandGaussianNoised(keys=["img"], prob=0.25, mean=0, std=0.1),
+ RandAdjustContrastd(keys=["img"], prob=0.25, gamma=(1, 2)),
+ RandGaussianSmoothd(keys=["img"], prob=0.25, sigma_x=(1, 2)),
+ RandHistogramShiftd(keys=["img"], prob=0.25, num_control_points=3),
+ RandGaussianSharpend(keys=["img"], prob=0.25),
+ EnsureTyped(keys=["img", "label"]),
+ ]
+)
+
+
+public_transforms = Compose(
+ [
+ CustomLoadImaged(keys=["img", "label"], image_only=True),
+ BoundaryExclusion(keys=["label"]),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img", "label"], channel_dim=-1),
+ RemoveRepeatedChanneld(keys=["label"], repeats=3), # label: (H, W)
+ ScaleIntensityd(keys=["img"], allow_missing_keys=True), # Do not scale label
+ # >>> Spatial transforms
+ SpatialPadd(keys=["img", "label"], spatial_size=512),
+ RandSpatialCropd(keys=["img", "label"], roi_size=512, random_size=False),
+ RandAxisFlipd(keys=["img", "label"], prob=0.5),
+ RandRotate90d(keys=["img", "label"], prob=0.5, spatial_axes=[0, 1]),
+ Rotate90d(k=1, keys=["label"], spatial_axes=(0, 1)),
+ Flipd(keys=["label"], spatial_axis=0),
+ EnsureTyped(keys=["img", "label"]),
+ ]
+)
+
+
+valid_transforms = Compose(
+ [
+ CustomLoadImaged(keys=["img", "label"], allow_missing_keys=True, image_only=True),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img", "label"], allow_missing_keys=True, channel_dim=-1),
+ RemoveRepeatedChanneld(keys=["label"], repeats=3),
+ ScaleIntensityd(keys=["img"], allow_missing_keys=True),
+ EnsureTyped(keys=["img", "label"], allow_missing_keys=True),
+ ]
+)
+
+tuning_transforms = Compose(
+ [
+ CustomLoadImaged(keys=["img"], image_only=True),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img"], channel_dim=-1),
+ ScaleIntensityd(keys=["img"]),
+ EnsureTyped(keys=["img"]),
+ ]
+)
+
+unlabeled_transforms = Compose(
+ [
+ # >>> Load and refine data --- img: (H, W, 3); label: (H, W)
+ CustomLoadImaged(keys=["img"], image_only=True),
+ CustomNormalizeImaged(
+ keys=["img"],
+ allow_missing_keys=True,
+ channel_wise=False,
+ percentiles=[0.0, 99.5],
+ ),
+ EnsureChannelFirstd(keys=["img"], channel_dim=-1),
+ RandZoomd(
+ keys=["img"],
+ prob=0.5,
+ min_zoom=0.25,
+ max_zoom=1.25,
+ mode=["area"],
+ keep_size=False,
+ ),
+ ScaleIntensityd(keys=["img"], allow_missing_keys=True), # Do not scale label
+ # >>> Spatial transforms
+ SpatialPadd(keys=["img"], spatial_size=512),
+ RandSpatialCropd(keys=["img"], roi_size=512, random_size=False),
+ EnsureTyped(keys=["img"]),
+ ]
+)
+
+
+def get_pred_transforms():
+ """Prediction preprocessing"""
+ pred_transforms = Compose(
+ [
+ # >>> Load and refine data
+ CustomLoadImage(image_only=True),
+ CustomNormalizeImage(channel_wise=False, percentiles=[0.0, 99.5]),
+ EnsureChannelFirst(channel_dim=-1), # image: (3, H, W)
+ ScaleIntensity(),
+ EnsureType(data_type="tensor"),
+ ]
+ )
+
+ return pred_transforms
diff --git a/annolid/segmentation/MEDIAR/train_tools/data_utils/utils.py b/annolid/segmentation/MEDIAR/train_tools/data_utils/utils.py
new file mode 100644
index 00000000..6e926a3c
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/data_utils/utils.py
@@ -0,0 +1,84 @@
+import os
+import json
+import numpy as np
+
+__all__ = ["split_train_valid", "path_decoder"]
+
+
+def split_train_valid(data_dicts, valid_portion=0.1):
+ """Split train/validata data according to the given proportion"""
+
+ train_dicts, valid_dicts = data_dicts, []
+ if valid_portion > 0:
+
+ # Obtain & shuffle data indices
+ num_data_dicts = len(data_dicts)
+ indices = np.arange(num_data_dicts)
+ np.random.shuffle(indices)
+
+ # Divide train/valid indices by the proportion
+ valid_size = int(num_data_dicts * valid_portion)
+ train_indices = indices[valid_size:]
+ valid_indices = indices[:valid_size]
+
+ # Assign data dicts by split indices
+ train_dicts = [data_dicts[idx] for idx in train_indices]
+ valid_dicts = [data_dicts[idx] for idx in valid_indices]
+
+ print(
+ "\n(DataLoaded) Training data size: %d, Validation data size: %d\n"
+ % (len(train_dicts), len(valid_dicts))
+ )
+
+ return train_dicts, valid_dicts
+
+
+def path_decoder(root, mapping_file, no_label=False, unlabeled=False):
+ """Decode img/label file paths from root & mapping directory.
+
+ Args:
+ root (str):
+ mapping_file (str): json file containing image & label file paths.
+ no_label (bool, optional): whether to include "label" key. Defaults to False.
+
+ Returns:
+ list: list of dictionary. (ex. [{"img": img_path, "label": label_path}, ...])
+ """
+
+ data_dicts = []
+
+ with open(mapping_file, "r") as file:
+ data = json.load(file)
+
+ for map_key in data.keys():
+
+ # If no_label, assign "img" key only
+ if no_label:
+ data_dict_item = [
+ {"img": os.path.join(root, elem["img"]),} for elem in data[map_key]
+ ]
+
+ # If label exists, assign both "img" and "label" keys
+ else:
+ data_dict_item = [
+ {
+ "img": os.path.join(root, elem["img"]),
+ "label": os.path.join(root, elem["label"]),
+ }
+ for elem in data[map_key]
+ ]
+
+ # Add refined datasets to be returned
+ data_dicts += data_dict_item
+
+ if unlabeled:
+ refined_data_dicts = []
+
+ # Exclude the corrupted image to prevent errror
+ for data_dict in data_dicts:
+ if "00504" not in data_dict["img"]:
+ refined_data_dicts.append(data_dict)
+
+ data_dicts = refined_data_dicts
+
+ return data_dicts
diff --git a/annolid/segmentation/MEDIAR/train_tools/measures.py b/annolid/segmentation/MEDIAR/train_tools/measures.py
new file mode 100644
index 00000000..b17509a4
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/measures.py
@@ -0,0 +1,200 @@
+"""
+Adapted from the following references:
+[1] https://github.com/JunMa11/NeurIPS-CellSeg/blob/main/baseline/compute_metric.py
+[2] https://github.com/stardist/stardist/blob/master/stardist/matching.py
+
+"""
+
+import numpy as np
+from skimage import segmentation
+from scipy.optimize import linear_sum_assignment
+from numba import jit
+
+__all__ = ["evaluate_f1_score_cellseg", "evaluate_f1_score"]
+
+
+def evaluate_f1_score_cellseg(masks_true, masks_pred, threshold=0.5):
+ """
+ Get confusion elements for cell segmentation results.
+ Boundary pixels are not considered during evaluation.
+ """
+
+ if np.prod(masks_true.shape) < (5000 * 5000):
+ masks_true = _remove_boundary_cells(masks_true.astype(np.int32))
+ masks_pred = _remove_boundary_cells(masks_pred.astype(np.int32))
+
+ tp, fp, fn = get_confusion(masks_true, masks_pred, threshold)
+
+ # Compute by Patch-based way for large images
+ else:
+ H, W = masks_true.shape
+ roi_size = 2000
+
+ # Get patch grid by roi_size
+ if H % roi_size != 0:
+ n_H = H // roi_size + 1
+ new_H = roi_size * n_H
+ else:
+ n_H = H // roi_size
+ new_H = H
+
+ if W % roi_size != 0:
+ n_W = W // roi_size + 1
+ new_W = roi_size * n_W
+ else:
+ n_W = W // roi_size
+ new_W = W
+
+ # Allocate values on the grid
+ gt_pad = np.zeros((new_H, new_W), dtype=masks_true.dtype)
+ pred_pad = np.zeros((new_H, new_W), dtype=masks_true.dtype)
+ gt_pad[:H, :W] = masks_true
+ pred_pad[:H, :W] = masks_pred
+
+ tp, fp, fn = 0, 0, 0
+
+ # Calculate confusion elements for each patch
+ for i in range(n_H):
+ for j in range(n_W):
+ gt_roi = _remove_boundary_cells(
+ gt_pad[
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ]
+ )
+ pred_roi = _remove_boundary_cells(
+ pred_pad[
+ roi_size * i : roi_size * (i + 1),
+ roi_size * j : roi_size * (j + 1),
+ ]
+ )
+ tp_i, fp_i, fn_i = get_confusion(gt_roi, pred_roi, threshold)
+ tp += tp_i
+ fp += fp_i
+ fn += fn_i
+
+ # Calculate f1 score
+ precision, recall, f1_score = evaluate_f1_score(tp, fp, fn)
+
+ return precision, recall, f1_score
+
+
+def evaluate_f1_score(tp, fp, fn):
+ """Evaluate F1-score for the given confusion elements"""
+
+ # Do not Compute on trivial results
+ if tp == 0:
+ precision, recall, f1_score = 0, 0, 0
+
+ else:
+ precision = tp / (tp + fp)
+ recall = tp / (tp + fn)
+ f1_score = 2 * (precision * recall) / (precision + recall)
+
+ return precision, recall, f1_score
+
+
+def _remove_boundary_cells(mask):
+ """Remove cells on the boundary from the mask"""
+
+ # Identify boundary cells
+ W, H = mask.shape
+ bd = np.ones((W, H))
+ bd[2 : W - 2, 2 : H - 2] = 0
+ bd_cells = np.unique(mask * bd)
+
+ # Remove cells on the boundary
+ for i in bd_cells[1:]:
+ mask[mask == i] = 0
+
+ # Allocate labels as sequential manner
+ new_label, _, _ = segmentation.relabel_sequential(mask)
+
+ return new_label
+
+
+def get_confusion(masks_true, masks_pred, threshold=0.5):
+ """Calculate confusion matrix elements: (TP, FP, FN)"""
+ num_gt_instances = np.max(masks_true)
+ num_pred_instances = np.max(masks_pred)
+
+ if num_pred_instances == 0:
+ print("No segmentation results!")
+ tp, fp, fn = 0, 0, 0
+
+ else:
+ # Calculate IoU and exclude background label (0)
+ iou = _get_iou(masks_true, masks_pred)
+ iou = iou[1:, 1:]
+
+ # Calculate true positives
+ tp = _get_true_positive(iou, threshold)
+ fp = num_pred_instances - tp
+ fn = num_gt_instances - tp
+
+ return tp, fp, fn
+
+
+def _get_true_positive(iou, threshold=0.5):
+ """Get true positive (TP) pixels at the given threshold"""
+
+ # Number of instances to be matched
+ num_matched = min(iou.shape[0], iou.shape[1])
+
+ # Find optimal matching by using IoU as tie-breaker
+ costs = -(iou >= threshold).astype(np.float) - iou / (2 * num_matched)
+ matched_gt_label, matched_pred_label = linear_sum_assignment(costs)
+
+ # Consider as the same instance only if the IoU is above the threshold
+ match_ok = iou[matched_gt_label, matched_pred_label] >= threshold
+ tp = match_ok.sum()
+
+ return tp
+
+
+def _get_iou(masks_true, masks_pred):
+ """Get the iou between masks_true and masks_pred"""
+
+ # Get overlap matrix (GT Instances Num, Pred Instance Num)
+ overlap = _label_overlap(masks_true, masks_pred)
+
+ # Predicted instance pixels
+ n_pixels_pred = np.sum(overlap, axis=0, keepdims=True)
+
+ # GT instance pixels
+ n_pixels_true = np.sum(overlap, axis=1, keepdims=True)
+
+ # Calculate intersection of union (IoU)
+ union = n_pixels_pred + n_pixels_true - overlap
+ iou = overlap / union
+
+ # Ensure numerical values
+ iou[np.isnan(iou)] = 0.0
+
+ return iou
+
+
+@jit(nopython=True)
+def _label_overlap(x, y):
+ """Get pixel overlaps between two masks
+
+ Parameters
+ ------------
+ x, y (np array; dtype int): 0=NO masks; 1,2... are mask labels
+
+ Returns
+ ------------
+ overlap (np array; dtype int): Overlaps of size [x.max()+1, y.max()+1]
+ """
+
+ # Make as 1D array
+ x, y = x.ravel(), y.ravel()
+
+ # Preallocate a Contact Map matrix
+ overlap = np.zeros((1 + x.max(), 1 + y.max()), dtype=np.uint)
+
+ # Calculate the number of shared pixels for each label
+ for i in range(len(x)):
+ overlap[x[i], y[i]] += 1
+
+ return overlap
diff --git a/annolid/segmentation/MEDIAR/train_tools/models/MEDIARFormer.py b/annolid/segmentation/MEDIAR/train_tools/models/MEDIARFormer.py
new file mode 100644
index 00000000..f7f19f2b
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/models/MEDIARFormer.py
@@ -0,0 +1,102 @@
+import torch
+import torch.nn as nn
+
+from segmentation_models_pytorch import MAnet
+from segmentation_models_pytorch.base.modules import Activation
+
+__all__ = ["MEDIARFormer"]
+
+
+class MEDIARFormer(MAnet):
+ """MEDIAR-Former Model"""
+
+ def __init__(
+ self,
+ encoder_name="mit_b5", # Default encoder
+ encoder_weights="imagenet", # Pre-trained weights
+ decoder_channels=(1024, 512, 256, 128, 64), # Decoder configuration
+ decoder_pab_channels=256, # Decoder Pyramid Attention Block channels
+ in_channels=3, # Number of input channels
+ classes=3, # Number of output classes
+ ):
+ # Initialize the MAnet model with provided parameters
+ super().__init__(
+ encoder_name=encoder_name,
+ encoder_weights=encoder_weights,
+ decoder_channels=decoder_channels,
+ decoder_pab_channels=decoder_pab_channels,
+ in_channels=in_channels,
+ classes=classes,
+ )
+
+ # Remove the default segmentation head as it's not used in this architecture
+ self.segmentation_head = None
+
+ # Modify all activation functions in the encoder and decoder from ReLU to Mish
+ _convert_activations(self.encoder, nn.ReLU, nn.Mish(inplace=True))
+ _convert_activations(self.decoder, nn.ReLU, nn.Mish(inplace=True))
+
+ # Add custom segmentation heads for different segmentation tasks
+ self.cellprob_head = DeepSegmentationHead(
+ in_channels=decoder_channels[-1], out_channels=1
+ )
+ self.gradflow_head = DeepSegmentationHead(
+ in_channels=decoder_channels[-1], out_channels=2
+ )
+
+ def forward(self, x):
+ """Forward pass through the network"""
+ # Ensure the input shape is correct
+ self.check_input_shape(x)
+
+ # Encode the input and then decode it
+ features = self.encoder(x)
+ decoder_output = self.decoder(*features)
+
+ # Generate masks for cell probability and gradient flows
+ cellprob_mask = self.cellprob_head(decoder_output)
+ gradflow_mask = self.gradflow_head(decoder_output)
+
+ # Concatenate the masks for output
+ masks = torch.cat([gradflow_mask, cellprob_mask], dim=1)
+
+ return masks
+
+
+class DeepSegmentationHead(nn.Sequential):
+ """Custom segmentation head for generating specific masks"""
+
+ def __init__(
+ self, in_channels, out_channels, kernel_size=3, activation=None, upsampling=1
+ ):
+ # Define a sequence of layers for the segmentation head
+ layers = [
+ nn.Conv2d(
+ in_channels,
+ in_channels // 2,
+ kernel_size=kernel_size,
+ padding=kernel_size // 2,
+ ),
+ nn.Mish(inplace=True),
+ nn.BatchNorm2d(in_channels // 2),
+ nn.Conv2d(
+ in_channels // 2,
+ out_channels,
+ kernel_size=kernel_size,
+ padding=kernel_size // 2,
+ ),
+ nn.UpsamplingBilinear2d(scale_factor=upsampling)
+ if upsampling > 1
+ else nn.Identity(),
+ Activation(activation) if activation else nn.Identity(),
+ ]
+ super().__init__(*layers)
+
+
+def _convert_activations(module, from_activation, to_activation):
+ """Recursively convert activation functions in a module"""
+ for name, child in module.named_children():
+ if isinstance(child, from_activation):
+ setattr(module, name, to_activation)
+ else:
+ _convert_activations(child, from_activation, to_activation)
diff --git a/annolid/segmentation/MEDIAR/train_tools/models/__init__.py b/annolid/segmentation/MEDIAR/train_tools/models/__init__.py
new file mode 100644
index 00000000..cc8bcdb6
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/models/__init__.py
@@ -0,0 +1 @@
+from .MEDIARFormer import *
diff --git a/annolid/segmentation/MEDIAR/train_tools/utils.py b/annolid/segmentation/MEDIAR/train_tools/utils.py
new file mode 100644
index 00000000..73b704df
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/train_tools/utils.py
@@ -0,0 +1,70 @@
+import torch
+import numpy as np
+import os, json, random
+from pprint import pprint
+
+__all__ = ["ConfLoader", "directory_setter", "random_seeder", "pprint_config"]
+
+
+class ConfLoader:
+ """
+ Load json config file using DictWithAttributeAccess object_hook.
+ ConfLoader(conf_name).opt attribute is the result of loading json config file.
+ """
+
+ class DictWithAttributeAccess(dict):
+ """
+ This inner class makes dict to be accessed same as class attribute.
+ For example, you can use opt.key instead of the opt['key'].
+ """
+
+ def __getattr__(self, key):
+ return self[key]
+
+ def __setattr__(self, key, value):
+ self[key] = value
+
+ def __init__(self, conf_name):
+ self.conf_name = conf_name
+ self.opt = self.__get_opt()
+
+ def __load_conf(self):
+ with open(self.conf_name, "r") as conf:
+ opt = json.load(
+ conf, object_hook=lambda dict: self.DictWithAttributeAccess(dict)
+ )
+ return opt
+
+ def __get_opt(self):
+ opt = self.__load_conf()
+ opt = self.DictWithAttributeAccess(opt)
+
+ return opt
+
+
+def directory_setter(path="./results", make_dir=False):
+ """
+ Make dictionary if not exists.
+ """
+ if not os.path.exists(path) and make_dir:
+ os.makedirs(path) # make dir if not exist
+ print("directory %s is created" % path)
+
+ if not os.path.isdir(path):
+ raise NotADirectoryError(
+ "%s is not valid. set make_dir=True to make dir." % path
+ )
+
+
+def random_seeder(seed):
+ """Fix randomness."""
+ torch.manual_seed(seed)
+ np.random.seed(seed)
+ random.seed(seed)
+ torch.backends.cudnn.deterministic = True
+ torch.backends.cudnn.benchmark = False
+
+def pprint_config(opt):
+ print("\n" + "=" * 50 + " Configuration " + "=" * 50)
+ pprint(opt, compact=True)
+ print("=" * 115 + "\n")
\ No newline at end of file
diff --git a/annolid/segmentation/MEDIAR/viz.py b/annolid/segmentation/MEDIAR/viz.py
new file mode 100644
index 00000000..7b39fe43
--- /dev/null
+++ b/annolid/segmentation/MEDIAR/viz.py
@@ -0,0 +1,29 @@
+import os
+from skimage import io
+import numpy as np
+
+def show_and_count_cells(output_path, img_name="img2_label.tiff", cmap="cividis"):
+ pred_path = os.path.join(output_path, img_name)
+
+ try:
+ pred = io.imread(pred_path)
+ except Exception as e:
+ print(f"Error loading image: {e}")
+ return
+
+ if pred is None:
+ print("Image is empty or could not be loaded.")
+ return
+
+ try:
+ io.imshow(pred, cmap=cmap)
+ io.show()
+ except Exception as e:
+ print(f"Error displaying image: {e}")
+
+ cell_count = len(np.unique(pred)) - 1 # Exclude the background
+ print(f"\n{cell_count} Cells detected!")
+
+# Example usage:
+output_path = "./results/mediar_base_prediction"
+show_and_count_cells(output_path,'OpenTest_004_label.tiff')
\ No newline at end of file
diff --git a/setup.py b/setup.py
index 0b32bb99..9f23c651 100644
--- a/setup.py
+++ b/setup.py
@@ -47,6 +47,13 @@
'Pillow>=9.3.0,<=9.5.0',
"chardet>=5.2.0",
"scikit-learn-extra >= 0.3.0",
+ 'monai>=1.3.0',
+ 'segmentation-models-pytorch>=0.3.3',
+ 'fastremap>=1.14.1',
+ 'numba>=0.57.1',
+ 'tifffile>=2023.4.12',
+ 'wandb>=0.16.2',
+ 'scikit-image',
# "segment-anything @ git+https://github.com/facebookresearch/segment-anything.git",
"segment-anything @ git+https://github.com/SysCV/sam-hq.git",
],
 +
+MEIDAR is a framework for efficient cell instance segmentation of multi-modality microscopy images. The above figure illustrates an overview of our approach. MEDIAR harmonizes data-centric and model-centric approaches as the learning and inference strategies, achieving a **0.9067** Mean F1-score on the validation datasets. We provide a brief description of methods that combined in the MEDIAR. Please refer to our paper for more information.
+# 2. Methods
+
+## **Data-Centric**
+- **Cell Aware Augmentation** : We apply two novel cell-aware augmentations. *cell-wisely intensity is randomization* (Cell Intensity Diversification) and *cell-wise boundary pixels exclusion* in the label. The boundary exclusion is adopted only in the pre-training phase.
+
+- **Two-phase Pretraining and Fine-tuning** : To extract knowledge from large public datasets, we first pretrained our model on public sets, then fine-tune.
+ - Pretraining : We use 7,2412 labeled images from four public datasets for pretraining: OmniPose, CellPose, LiveCell and DataScienceBowl-2018. MEDIAR takes two different phases for the pretraining. the MEDIAR-Former model with encoder parameters initialized from ImageNet-1k pretraining.
+
+ - Fine-tuning : We use two different model for ensemble. First model is fine-tuned 200 epochs using target datasets. Second model is fine-tuned 25 epochs using both target and public datsets.
+
+- **Modality Discovery & Amplified Sampling** : To balance towards the latent modalities in the datasets, we conduct K-means clustering and discover 40 modalities. In the training phase, we over-sample the minor cluster samples.
+
+- **Cell Memory Replay** : We concatenate the data from the public dataset with a small portion to the batch and train with boundary-excluded labels.
+
+## **Model-Centric**
+- **MEDIAR-Former Architecture** : MEDIAR-Former follows the design paradigm of U-Net, but use SegFormer and MA-Net for the encoder and decoder. The two heads of MEDIAR-Former predicts cell probability and gradieng flow.
+
+
+
+MEIDAR is a framework for efficient cell instance segmentation of multi-modality microscopy images. The above figure illustrates an overview of our approach. MEDIAR harmonizes data-centric and model-centric approaches as the learning and inference strategies, achieving a **0.9067** Mean F1-score on the validation datasets. We provide a brief description of methods that combined in the MEDIAR. Please refer to our paper for more information.
+# 2. Methods
+
+## **Data-Centric**
+- **Cell Aware Augmentation** : We apply two novel cell-aware augmentations. *cell-wisely intensity is randomization* (Cell Intensity Diversification) and *cell-wise boundary pixels exclusion* in the label. The boundary exclusion is adopted only in the pre-training phase.
+
+- **Two-phase Pretraining and Fine-tuning** : To extract knowledge from large public datasets, we first pretrained our model on public sets, then fine-tune.
+ - Pretraining : We use 7,2412 labeled images from four public datasets for pretraining: OmniPose, CellPose, LiveCell and DataScienceBowl-2018. MEDIAR takes two different phases for the pretraining. the MEDIAR-Former model with encoder parameters initialized from ImageNet-1k pretraining.
+
+ - Fine-tuning : We use two different model for ensemble. First model is fine-tuned 200 epochs using target datasets. Second model is fine-tuned 25 epochs using both target and public datsets.
+
+- **Modality Discovery & Amplified Sampling** : To balance towards the latent modalities in the datasets, we conduct K-means clustering and discover 40 modalities. In the training phase, we over-sample the minor cluster samples.
+
+- **Cell Memory Replay** : We concatenate the data from the public dataset with a small portion to the batch and train with boundary-excluded labels.
+
+## **Model-Centric**
+- **MEDIAR-Former Architecture** : MEDIAR-Former follows the design paradigm of U-Net, but use SegFormer and MA-Net for the encoder and decoder. The two heads of MEDIAR-Former predicts cell probability and gradieng flow.
+
+ +
+- **Gradient Flow Tracking** : We utilize gradient flow tracking proposed by [CellPose](https://github.com/MouseLand/cellpose).
+
+- **Ensemble with Stochastic TTA**: During the inference, the MEIDAR conduct prediction as sliding-window manner with importance map generated by the gaussian filter. We use two fine-tuned models from phase1 and phase2 pretraining, and ensemble their outputs by summation. For each outputs, test-time augmentation is used.
+# 3. Experiments
+
+### **Dataset**
+- Official Dataset
+ - We are provided the target dataset from [Weakly Supervised Cell Segmentation in Multi-modality High-Resolution Microscopy Images](https://neurips22-cellseg.grand-challenge.org/). It consists of 1,000 labeled images, 1,712 unlabeled images and 13 unlabeled whole slide image from various microscopy types, tissue types, and staining types. Validation set is given with 101 images including 1 whole slide image.
+
+- Public Dataset
+ - [OmniPose](http://www.cellpose.org/dataset_omnipose) : contains mixtures of 14 bacterial species. We only use 611 bacterial cell microscopy images and discard 118 worm images.
+ - [CellPose](https://www.cellpose.org/dataset) : includes Cytoplasm, cellular microscopy, fluorescent cells images. We used 551 images by discarding 58 non-microscopy images. We convert all images as gray-scale.
+ - [LiveCell](https://github.com/sartorius-research/LIVECell) : is a large-scale dataset with 5,239 images containing 1,686,352 individual cells annotated by trained crowdsources from 8 distinct cell types.
+ - [DataScienceBowl 2018](https://www.kaggle.com/competitions/sartorius-cell-instance-segmentation/overview) : 841 images contain 37,333 cells from 22 cell types, 15 image resolutions, and five visually similar groups.
+
+### **Testing steps**
+- **Ensemble Prediction with TTA** : MEDIAR uses sliding-window inference with the overlap size between the adjacent patches as 0.6 and gaussian importance map. To predict the different views on the image, MEDIAR uses Test-Time Augmentation (TTA) for the model prediction and ensemble two models described in **Two-phase Pretraining and Fine-tuning**.
+
+- **Inference time** : MEDIAR conducts most images in less than 1sec and it depends on the image size and the number of cells, even with ensemble prediction with TTA. Detailed evaluation-time results are in the paper.
+
+### **Preprocessing & Augmentations**
+| Strategy | Type | Probability |
+|----------|:-------------|------|
+| `Clip` | Pre-processing | . |
+| `Normalization` | Pre-processing | . |
+| `Scale Intensity` | Pre-processing | . |
+| `Zoom` | Spatial Augmentation | 0.5 |
+| `Spatial Crop` | Spatial Augmentation | 1.0 |
+| `Axis Flip` | Spatial Augmentation | 0.5 |
+| `Rotation` | Spatial Augmentation | 0.5 |
+| `Cell-Aware Intensity` | Intensity Augmentation | 0.25 |
+| `Gaussian Noise` | Intensity Augmentation | 0.25 |
+| `Contrast Adjustment` | Intensity Augmentation | 0.25 |
+| `Gaussian Smoothing` | Intensity Augmentation | 0.25 |
+| `Histogram Shift` | Intensity Augmentation | 0.25 |
+| `Gaussian Sharpening` | Intensity Augmentation | 0.25 |
+| `Boundary Exclusion` | Others | . |
+
+
+| Learning Setups | Pretraining | Fine-tuning |
+|----------------------------------------------------------------------|---------------------------------------------------------|---------------------------------------------------------|
+| Initialization (Encoder) | Imagenet-1k pretrained | from Pretraining |
+| Initialization (Decoder, Head) | He normal initialization | from Pretraining|
+| Batch size | 9 | 9 |
+| Total epochs | 80 (60) | 200 (25) |
+| Optimizer | AdamW | AdamW |
+| Initial learning rate (lr) | 5e-5 | 2e-5 |
+| Lr decay schedule | Cosine scheduler (100 interval) | Cosine scheduler (100 interval) |
+| Loss function | MSE, BCE | MSE, BCE |
+
+# 4. Results
+### **Validation Dataset**
+- Quantitative Evaluation
+ - Our MEDIAR achieved **0.9067** validation mean F1-score.
+- Qualitative Evaluation
+
+
+- **Gradient Flow Tracking** : We utilize gradient flow tracking proposed by [CellPose](https://github.com/MouseLand/cellpose).
+
+- **Ensemble with Stochastic TTA**: During the inference, the MEIDAR conduct prediction as sliding-window manner with importance map generated by the gaussian filter. We use two fine-tuned models from phase1 and phase2 pretraining, and ensemble their outputs by summation. For each outputs, test-time augmentation is used.
+# 3. Experiments
+
+### **Dataset**
+- Official Dataset
+ - We are provided the target dataset from [Weakly Supervised Cell Segmentation in Multi-modality High-Resolution Microscopy Images](https://neurips22-cellseg.grand-challenge.org/). It consists of 1,000 labeled images, 1,712 unlabeled images and 13 unlabeled whole slide image from various microscopy types, tissue types, and staining types. Validation set is given with 101 images including 1 whole slide image.
+
+- Public Dataset
+ - [OmniPose](http://www.cellpose.org/dataset_omnipose) : contains mixtures of 14 bacterial species. We only use 611 bacterial cell microscopy images and discard 118 worm images.
+ - [CellPose](https://www.cellpose.org/dataset) : includes Cytoplasm, cellular microscopy, fluorescent cells images. We used 551 images by discarding 58 non-microscopy images. We convert all images as gray-scale.
+ - [LiveCell](https://github.com/sartorius-research/LIVECell) : is a large-scale dataset with 5,239 images containing 1,686,352 individual cells annotated by trained crowdsources from 8 distinct cell types.
+ - [DataScienceBowl 2018](https://www.kaggle.com/competitions/sartorius-cell-instance-segmentation/overview) : 841 images contain 37,333 cells from 22 cell types, 15 image resolutions, and five visually similar groups.
+
+### **Testing steps**
+- **Ensemble Prediction with TTA** : MEDIAR uses sliding-window inference with the overlap size between the adjacent patches as 0.6 and gaussian importance map. To predict the different views on the image, MEDIAR uses Test-Time Augmentation (TTA) for the model prediction and ensemble two models described in **Two-phase Pretraining and Fine-tuning**.
+
+- **Inference time** : MEDIAR conducts most images in less than 1sec and it depends on the image size and the number of cells, even with ensemble prediction with TTA. Detailed evaluation-time results are in the paper.
+
+### **Preprocessing & Augmentations**
+| Strategy | Type | Probability |
+|----------|:-------------|------|
+| `Clip` | Pre-processing | . |
+| `Normalization` | Pre-processing | . |
+| `Scale Intensity` | Pre-processing | . |
+| `Zoom` | Spatial Augmentation | 0.5 |
+| `Spatial Crop` | Spatial Augmentation | 1.0 |
+| `Axis Flip` | Spatial Augmentation | 0.5 |
+| `Rotation` | Spatial Augmentation | 0.5 |
+| `Cell-Aware Intensity` | Intensity Augmentation | 0.25 |
+| `Gaussian Noise` | Intensity Augmentation | 0.25 |
+| `Contrast Adjustment` | Intensity Augmentation | 0.25 |
+| `Gaussian Smoothing` | Intensity Augmentation | 0.25 |
+| `Histogram Shift` | Intensity Augmentation | 0.25 |
+| `Gaussian Sharpening` | Intensity Augmentation | 0.25 |
+| `Boundary Exclusion` | Others | . |
+
+
+| Learning Setups | Pretraining | Fine-tuning |
+|----------------------------------------------------------------------|---------------------------------------------------------|---------------------------------------------------------|
+| Initialization (Encoder) | Imagenet-1k pretrained | from Pretraining |
+| Initialization (Decoder, Head) | He normal initialization | from Pretraining|
+| Batch size | 9 | 9 |
+| Total epochs | 80 (60) | 200 (25) |
+| Optimizer | AdamW | AdamW |
+| Initial learning rate (lr) | 5e-5 | 2e-5 |
+| Lr decay schedule | Cosine scheduler (100 interval) | Cosine scheduler (100 interval) |
+| Loss function | MSE, BCE | MSE, BCE |
+
+# 4. Results
+### **Validation Dataset**
+- Quantitative Evaluation
+ - Our MEDIAR achieved **0.9067** validation mean F1-score.
+- Qualitative Evaluation
+ +
+- Failure Cases
+
+
+- Failure Cases
+ +
+### **Test Dataset**
+
+
+
+
+# 5. Reproducing
+
+### **Our Environment**
+| Computing Infrastructure| |
+|-------------------------|----------------------------------------------------------------------|
+| System | Ubuntu 18.04.5 LTS |
+| CPU | AMD EPYC 7543 32-Core Processor CPU@2.26GHz |
+| RAM | 500GB; 3.125MT/s |
+| GPU (number and type) | NVIDIA A5000 (24GB) 2ea |
+| CUDA version | 11.7 |
+| Programming language | Python 3.9 |
+| Deep learning framework | Pytorch (v1.12, with torchvision v0.13.1) |
+| Code dependencies | MONAI (v0.9.0), Segmentation Models (v0.3.0) |
+| Specific dependencies | None |
+
+To install requirements:
+
+```
+pip install -r requirements.txt
+wandb off
+```
+
+## Dataset
+- The datasets directories under the root should the following structure:
+
+```
+ Root
+ ├── Datasets
+ │ ├── images (images can have various extensions: .tif, .tiff, .png, .bmp ...)
+ │ │ ├── cell_00001.png
+ │ │ ├── cell_00002.tif
+ │ │ ├── cell_00003.xxx
+ │ │ ├── ...
+ │ └── labels (labels must have .tiff extension.)
+ │ │ ├── cell_00001_label.tiff
+ │ │ ├── cell_00002.label.tiff
+ │ │ ├── cell_00003.label.tiff
+ │ │ ├── ...
+ └── ...
+```
+
+Before execute the codes, run the follwing code to generate path mappting json file:
+
+```python
+python ./generate_mapping.py --root=
+
+### **Test Dataset**
+
+
+
+
+# 5. Reproducing
+
+### **Our Environment**
+| Computing Infrastructure| |
+|-------------------------|----------------------------------------------------------------------|
+| System | Ubuntu 18.04.5 LTS |
+| CPU | AMD EPYC 7543 32-Core Processor CPU@2.26GHz |
+| RAM | 500GB; 3.125MT/s |
+| GPU (number and type) | NVIDIA A5000 (24GB) 2ea |
+| CUDA version | 11.7 |
+| Programming language | Python 3.9 |
+| Deep learning framework | Pytorch (v1.12, with torchvision v0.13.1) |
+| Code dependencies | MONAI (v0.9.0), Segmentation Models (v0.3.0) |
+| Specific dependencies | None |
+
+To install requirements:
+
+```
+pip install -r requirements.txt
+wandb off
+```
+
+## Dataset
+- The datasets directories under the root should the following structure:
+
+```
+ Root
+ ├── Datasets
+ │ ├── images (images can have various extensions: .tif, .tiff, .png, .bmp ...)
+ │ │ ├── cell_00001.png
+ │ │ ├── cell_00002.tif
+ │ │ ├── cell_00003.xxx
+ │ │ ├── ...
+ │ └── labels (labels must have .tiff extension.)
+ │ │ ├── cell_00001_label.tiff
+ │ │ ├── cell_00002.label.tiff
+ │ │ ├── cell_00003.label.tiff
+ │ │ ├── ...
+ └── ...
+```
+
+Before execute the codes, run the follwing code to generate path mappting json file:
+
+```python
+python ./generate_mapping.py --root=