diff --git a/README.md b/README.md
index 991bb436..4be48328 100644
--- a/README.md
+++ b/README.md
@@ -10,10 +10,10 @@
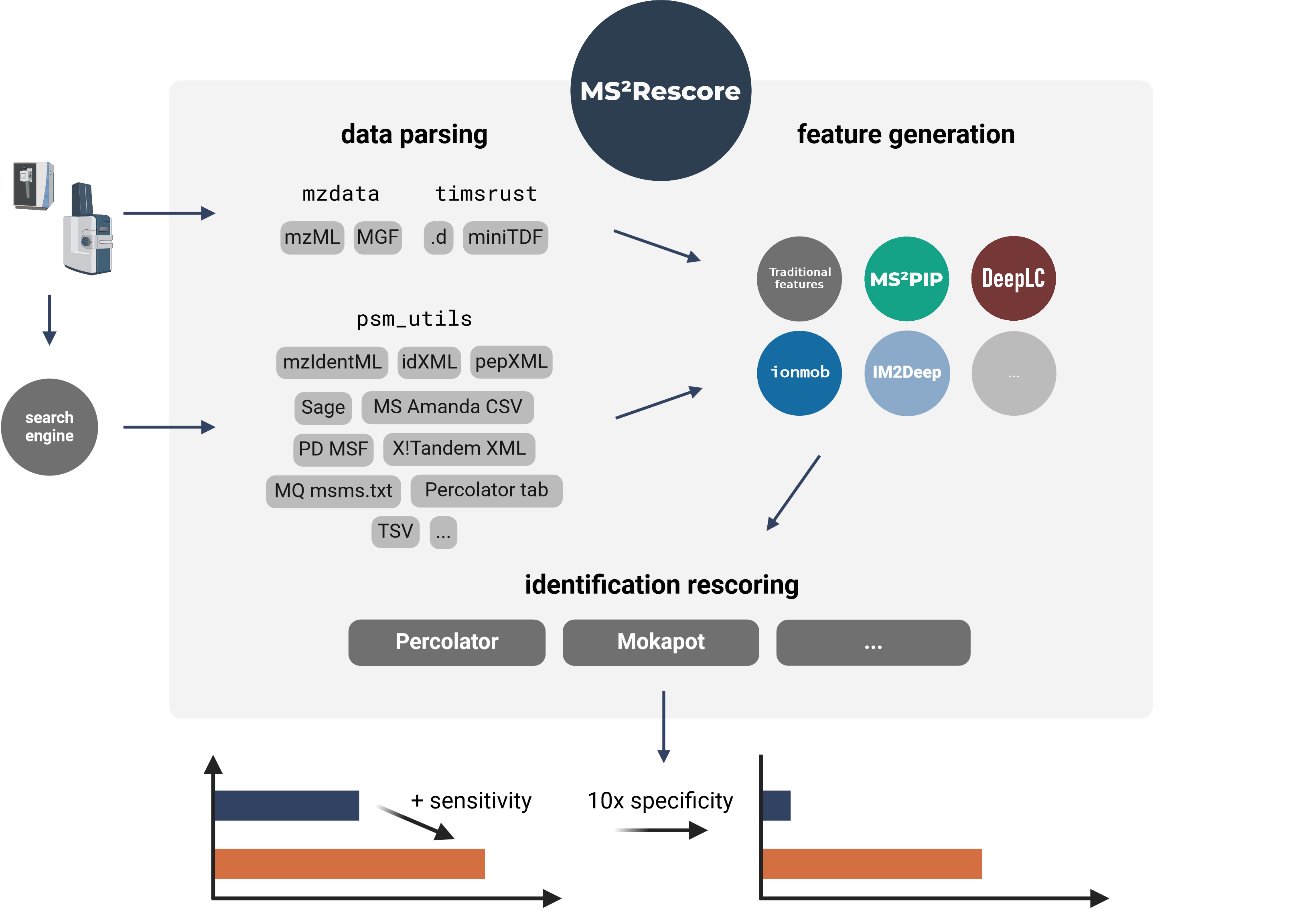
Modular and user-friendly platform for AI-assisted rescoring of peptide identifications
-> ⚠️ Note: This is the documentation for the fully redeveloped version 3.0 of MS²Rescore, which is
-> now in the beta stage. While MS²Rescore 3.0 has been drastically improved over the previous
-> version, you might run into some unforeseen issues. Please report any issues you encounter on the
-> [issue tracker][issues] or post your questions on the [GitHub Discussions][discussions] forum.
+> ⚠️ Note: This is the documentation for the fully redeveloped version 3.0 of MS²Rescore. While
+> MS²Rescore 3.0 has been drastically improved over the previous version, you might run into some
+> unforeseen issues. Please report any issues you encounter on the [issue tracker][issues] or post
+> your questions on the [GitHub Discussions][discussions] forum.
## About MS²Rescore
@@ -25,6 +25,8 @@ identifications, which allows you to get **more peptide IDs** at the same false
number of peptide IDs. MS²Rescore is **ideal for challenging proteomics identification workflows**,
such as proteogenomics, metaproteomics, or immunopeptidomics.
+
+
MS²Rescore can read peptide identifications in any format supported by [psm_utils][psm_utils]
(see [Supported file formats][file-formats]) and has been tested with various search engines output
files:
@@ -32,6 +34,7 @@ files:
- [MS Amanda](http://ms.imp.ac.at/?goto=msamanda) `.csv`
- [Sage](https://github.com/lazear/sage) `.sage.tsv`
- [PeptideShaker](https://compomics.github.io/projects/peptide-shaker.html) `.mzid`
+- [ProteomeDiscoverer](#)`.msf`
- [MSGFPlus](https://omics.pnl.gov/software/ms-gf) `.mzid`
- [Mascot](https://www.matrixscience.com/) `.mzid`
- [MaxQuant](https://www.maxquant.org/) `msms.txt`
@@ -45,6 +48,12 @@ MS²Rescore is available as a [desktop application][desktop], a [command line to
**Latest MS²Rescore publication:**
+> **MS²Rescore 3.0 is a modular, flexible, and user-friendly platform to boost peptide identifications, as showcased with MS Amanda 3.0.**
+> Louise Marie Buur*, Arthur Declercq*, Marina Strobl, Robbin Bouwmeester, Sven Degroeve, Lennart Martens, Viktoria Dorfer*, and Ralf Gabriels*.
+> _ChemRxiv_ (2023) [doi:10.26434/chemrxiv-2023-rvr9n](https://doi.org/10.26434/chemrxiv-2023-rvr9n)
\*contributed equally
+
+**MS²Rescore for immunopeptidomics:**
+
> **MS2Rescore: Data-driven rescoring dramatically boosts immunopeptide identification rates.**
> Arthur Declercq, Robbin Bouwmeester, Aurélie Hirschler, Christine Carapito, Sven Degroeve, Lennart Martens, and Ralf Gabriels.
> _Molecular & Cellular Proteomics_ (2021) [doi:10.1016/j.mcpro.2022.100266](https://doi.org/10.1016/j.mcpro.2022.100266)
diff --git a/docs/source/_static/img/gui-example-xtandem-advanced.png b/docs/source/_static/img/gui-example-xtandem-advanced.png
new file mode 100644
index 00000000..9a69db8e
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-advanced.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-finished.png b/docs/source/_static/img/gui-example-xtandem-finished.png
new file mode 100644
index 00000000..62ce2b68
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-finished.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-modifications-before.png b/docs/source/_static/img/gui-example-xtandem-modifications-before.png
new file mode 100644
index 00000000..c43f2ad5
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-modifications-before.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-modifications-filled.png b/docs/source/_static/img/gui-example-xtandem-modifications-filled.png
new file mode 100644
index 00000000..8ec4e22e
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-modifications-filled.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-output-files.png b/docs/source/_static/img/gui-example-xtandem-output-files.png
new file mode 100644
index 00000000..38986a69
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-output-files.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-processes.png b/docs/source/_static/img/gui-example-xtandem-processes.png
new file mode 100644
index 00000000..0e1493ad
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-processes.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-progress.png b/docs/source/_static/img/gui-example-xtandem-progress.png
new file mode 100644
index 00000000..3a4e3642
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-progress.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-psm-file.png b/docs/source/_static/img/gui-example-xtandem-psm-file.png
new file mode 100644
index 00000000..bb91365b
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-psm-file.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-psm-filetype.png b/docs/source/_static/img/gui-example-xtandem-psm-filetype.png
new file mode 100644
index 00000000..194c258d
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-psm-filetype.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-spectra.png b/docs/source/_static/img/gui-example-xtandem-spectra.png
new file mode 100644
index 00000000..f8c28c70
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-spectra.png differ
diff --git a/docs/source/_static/img/gui-example-xtandem-start.png b/docs/source/_static/img/gui-example-xtandem-start.png
new file mode 100644
index 00000000..c5389fbc
Binary files /dev/null and b/docs/source/_static/img/gui-example-xtandem-start.png differ
diff --git a/docs/source/_static/img/gui-overview.png b/docs/source/_static/img/gui-overview.png
new file mode 100644
index 00000000..622c7f4f
Binary files /dev/null and b/docs/source/_static/img/gui-overview.png differ
diff --git a/docs/source/_static/img/gui-screenshot-old.png b/docs/source/_static/img/gui-screenshot-old.png
new file mode 100644
index 00000000..4b4fc665
Binary files /dev/null and b/docs/source/_static/img/gui-screenshot-old.png differ
diff --git a/docs/source/_static/img/gui-screenshot.png b/docs/source/_static/img/gui-screenshot.png
index 4b4fc665..fa49e22b 100644
Binary files a/docs/source/_static/img/gui-screenshot.png and b/docs/source/_static/img/gui-screenshot.png differ
diff --git a/docs/source/_static/img/ms2rescore-overview.png b/docs/source/_static/img/ms2rescore-overview.png
new file mode 100644
index 00000000..d4a4001d
Binary files /dev/null and b/docs/source/_static/img/ms2rescore-overview.png differ
diff --git a/docs/source/_static/img/percolator-install-path.png b/docs/source/_static/img/percolator-install-path.png
new file mode 100644
index 00000000..f26db208
Binary files /dev/null and b/docs/source/_static/img/percolator-install-path.png differ
diff --git a/docs/source/_static/img/qc-reports.png b/docs/source/_static/img/qc-reports.png
new file mode 100644
index 00000000..ab892cb5
Binary files /dev/null and b/docs/source/_static/img/qc-reports.png differ
diff --git a/docs/source/api/ms2rescore.feature_generators.rst b/docs/source/api/ms2rescore.feature_generators.rst
index 6a0b49f9..1a457ee6 100644
--- a/docs/source/api/ms2rescore.feature_generators.rst
+++ b/docs/source/api/ms2rescore.feature_generators.rst
@@ -35,10 +35,10 @@ ms2rescore.feature_generators.deeplc
-ms2rescore.feature_generators.ms2pip
+ms2rescore.feature_generators.ionmob
####################################
-.. automodule:: ms2rescore.feature_generators.ms2pip
+.. automodule:: ms2rescore.feature_generators.ionmob
:members:
@@ -48,3 +48,11 @@ ms2rescore.feature_generators.maxquant
.. automodule:: ms2rescore.feature_generators.maxquant
:members:
+
+
+
+ms2rescore.feature_generators.ms2pip
+####################################
+
+.. automodule:: ms2rescore.feature_generators.ms2pip
+ :members:
diff --git a/docs/source/gui.rst b/docs/source/gui.rst
index 13f14858..e818e05d 100644
--- a/docs/source/gui.rst
+++ b/docs/source/gui.rst
@@ -2,4 +2,202 @@
Graphical user interface
************************
-[TODO]
+
+Installation
+============
+
+The MS²Rescore desktop application can be installed on Windows with a
+:ref:`one-click installer `. Alternatively, or on other platforms, follow the
+:ref:`Python package installation instructions `.
+
+
+Starting the application
+========================
+
+If installed with the one-click installer, simply start MS²Rescore from the start menu or with the
+desktop shortcut. Otherwise, start the application from the
+:ref:`command line ` with the command ``ms2rescore-gui`` or with
+``python -m ms2rescore.gui``.
+
+
+Application overview
+====================
+
+The MS²Rescore graphical user interface is divided into three main sections:
+
+1. A side bar with references, window controls, and the current version number.
+2. The configuration pane with input file selection, and parameter configuration.
+3. The application log pane with the status output.
+
+On the bottom of the window, the application log level can be selected. The log level determines
+which messages are shown in the application log pane. On the bottom right, the application can be
+started with the "Start" button. The "Stop" button can be used to stop the application at any time
+during the execution.
+
+.. figure:: ../_static/img/gui-overview.png
+ :width: 100%
+ :alt: MS²Rescore graphical user interface
+
+ Overview of the MS²Rescore desktop application.
+
+
+Configuring MS²Rescore
+======================
+
+Input file selection
+^^^^^^^^^^^^^^^^^^^^
+
+The main input for MS²Rescore are the PSM file(s) (search engine output) and the spectrum file(s).
+See :ref:`Input files` for more information.
+
+One or more PSM files can be selected from the file system with the "Browse files" button under.
+To make ensure correct reading of the file, specify the file type with from the drop-down menu.
+
+.. figure:: ../_static/img/gui-example-xtandem-psm-file.png
+ :width: 60%
+ :alt: PSM file selection
+
+ PSM file selection
+
+
+.. figure:: ../_static/img/gui-example-xtandem-psm-filetype.png
+ :width: 60%
+ :alt: PSM file type selection
+
+ PSM file type selection
+
+
+To select a single spectrum file (mzML or MGF), click the "Browse files" button. To select a
+folder with spectrum files, click the "Browse directories" button.
+
+.. figure:: ../_static/img/gui-example-xtandem-spectra.png
+ :width: 60%
+ :alt: Spectrum file selection
+
+ Spectrum file selection
+
+
+Optionally, for protein inference information, a FASTA file can also be provided. Ensure that
+this file contains the same protein sequences as the search database used for the search engine.
+If a FASTA file is provided, protein digestion settings may need to be configured in the rescoring
+engine configuration.
+
+
+Number of processes
+^^^^^^^^^^^^^^^^^^^
+
+The number of processes can be configured to run the application in parallel. The default is to
+use all available CPU cores. The number of processes can be reduced to avoid overloading the
+system or to avoid memory issues. A number under 16 is recommended.
+
+
+Modification mapping
+^^^^^^^^^^^^^^^^^^^^
+
+Depending on the search engine, the peptide modification labels will have to be mapped
+to labels that can be understood by MS²Rescore. For example, X!Tandem uses mass shift labels, such
+as ``+57.02146`` for carbamidomethylation. However, tools such as DeepLC requires the atomic
+composition for all modifications. As this cannot be derived from the mass shift (or other labels
+that are not known to MS²Rescore), a mapping has to be provided.
+
+.. figure:: ../_static/img/gui-example-xtandem-modifications-before.png
+ :width: 70%
+ :alt: Modification mapping
+
+ Modification mapping configuration. Click the plus sign to add more rows.
+
+
+In modification mapping, click the plus sign to add more rows to the table, or click the minus sign
+to remove rows. In the first column "Search engine label", enter the modification label as it
+appears in the PSM file. In the second column "ProForma label", enter a ProForma-compatible
+modification label. More information on accepted labels can be found in :ref:`Parsing modification
+labels`.
+
+.. figure:: ../_static/img/gui-example-xtandem-modifications-filled.png
+ :width: 70%
+ :alt: Modification mapping
+
+ Modification mapping configuration for the X!Tandem example. Mass shift labels from X!Tandem
+ are mapped to ProForma UniMod labels.
+
+
+Fixed modifications
+^^^^^^^^^^^^^^^^^^^
+
+If the search engine PSM file does not contain information on which fixed modifications were used,
+this must be specified in the MS²Rescore configuration. At the time of writing, only MaxQuant
+``msms.txt``` files do not contain this information. For all other search engines, this information
+is contained in the PSM file and the following field can be left empty.
+
+
+Advanced options
+^^^^^^^^^^^^^^^^
+
+Most advanced options are only required for specific use cases or with specific search engine PSM
+files. All options are listed in the :doc:`userguide/configuration` section of the user guide.
+
+In the X!Tandem example, only the `PSM ID regex pattern` option is required. This option is used
+to extract the spectrum ID from the PSM file. The spectrum ID is used to match the PSM to the
+spectrum file. See :ref:`Mapping PSMs to spectra` for more information.
+
+.. figure:: ../_static/img/gui-example-xtandem-advanced.png
+ :width: 70%
+ :alt: Advanced options
+
+ Advanced options
+
+
+For reference, all parameters for the X!Tandem example are also listed in the example
+configuration file on
+`GitHub `_.
+
+
+Starting the rescoring process
+==============================
+
+After the configuration is complete, click the "Start" button to start the rescoring process.
+The application will show the progress in the application log pane. The log level can be changed
+before the run to show more or less information.
+
+.. figure:: ../_static/img/gui-example-xtandem-progress.png
+ :width: 100%
+ :alt: Running application
+
+ Running application with log output
+
+
+A pop up will appear when the application is finished, or when an error occurred. If an error
+has occurred, the error message in the pop up should provide some insight into what went wrong.
+If the error message is not clear, please report the issue on the
+`GitHub issue tracker `_ or post your question on
+the `Discussion forum `_.
+
+.. figure:: ../_static/img/gui-example-xtandem-finished.png
+ :width: 40%
+ :alt: Pop up when MS²Rescore is finished
+
+ Pop up when MS²Rescore is finished
+
+
+Viewing the results
+===================
+
+After a successful run, the output files can be found in the directory of the input PSM file, or
+in the specified output directory. The most important files are the ``*.ms2rescore.psms.tsv`` file,
+which contains all PSMs with their new scores, and the ``*.ms2rescore.report.html`` file, which
+contains interactive charts that visualize the results and various quality control metrics. See
+:ref:`Output files` for more information.
+
+.. figure:: ../_static/img/gui-example-xtandem-output-files.png
+ :width: 100%
+ :alt: Output files
+
+ Overview of the output files after rescoring the X!Tandem example.
+
+Double click the ``*.ms2rescore.report.html`` file to open it in the default web browser:
+
+.. figure:: ../_static/img/qc-reports.png
+ :width: 100%
+ :alt: Rescoring report
+
+ Rescoring QC report with interactive charts.
diff --git a/docs/source/installation.rst b/docs/source/installation.rst
index 74da2ff8..e196ae7c 100644
--- a/docs/source/installation.rst
+++ b/docs/source/installation.rst
@@ -48,7 +48,7 @@ Docker container
:target: https://quay.io/repository/biocontainers/ms2rescore
First check the latest version tag on
-`biocontainers/ms2rescore/tags `__.
+`biocontainers/ms2rescore/tags `_.
Then pull and run the container with:
.. code-block:: bash
@@ -60,6 +60,23 @@ files, ```` is the container version tag, and ```` ar
command line options (see :ref:`Command line interface`).
+Installing Percolator
+=====================
+
+To use :ref:`percolator` as rescoring engine, it must be installed separately. Percolator is
+available for most platforms and can be downloaded from the
+`GitHub releases page `_. Ensure that
+the ``percolator`` executable is in your ``PATH``. On Windows, this can be done by checking the
+``Add percolator to the system PATH for current user`` option during installation:
+
+.. figure:: ../_static/img/percolator-install-path.png
+ :width: 60%
+ :alt: Percolator installation on Windows
+
+.. note::
+ Alternatively, :ref:`mokapot` can be used as rescoring engine, which does not require a separate
+ installation.
+
For development
===============
diff --git a/docs/source/userguide/configuration.rst b/docs/source/userguide/configuration.rst
index 8eecff89..52036aa7 100644
--- a/docs/source/userguide/configuration.rst
+++ b/docs/source/userguide/configuration.rst
@@ -140,6 +140,10 @@ be configured separately. For instance:
:alt: fixed modifications configuration in GUI
+.. caution::
+ Most search engines DO return fixed modifications as part of the modified peptide sequences.
+ In these cases, they must NOT be added to the ``fixed_modifications`` configuration.
+
Mapping PSMs to spectra
=======================
diff --git a/docs/source/userguide/input-files.rst b/docs/source/userguide/input-files.rst
index 21f47744..08129b7f 100644
--- a/docs/source/userguide/input-files.rst
+++ b/docs/source/userguide/input-files.rst
@@ -2,24 +2,30 @@
Input files
###########
-PSM file
-========
+PSM file(s)
+===========
-[todo]
+The peptide-spectrum match (PSM) file is generally the output from a proteomics search engine.
+This file serves as the main input to MS²Rescore. One or multiple PSM files can be provided at
+once. Note that merging PSMs from different MS runs could have an impact on the correctness of
+the FDR control.
-As a general rule, MS²Rescore always needs access to all target and decoy PSMs, not
-only the FDR-filtered targets.
+Various PSM file types are supported. The type can be specified with the ``psm_file_type`` option.
+Check the list of :py:mod:`psm_utils` tags in the
+:external+psm_utils:ref:`supported file formats ` section. Depending on the
+file extension, the file type can also be inferred from the file name. In that case,
+``psm_file_type`` option can be set to ``infer``.
-The ``psm_file_type`` can be one of the :py:mod:`psm_utils` tags as listed in the
-:external+psm_utils:ref:`supported file formats `. Depending on the file
-extension, the file type can also be inferred from the file name. In that case, ``psm_file_type``
-option can be set to ``infer``.
+.. attention::
+ As a general rule, MS²Rescore always needs access to **all target and decoy PSMs, without any
+ FDR-filtering**. For some search engines, this means that the FDR-filter should be disabled or
+ set to 100%.
Spectrum file(s)
================
-[todo]
-
-If the ``spectrum_path`` is a directory, MS²Rescore will search for spectrum files in the
-directory according to the run names in the PSM file.
+Spectrum files are required for some feature generators. Both ``mzML`` and ``mgf`` formats are
+supported. The ``spectrum_path`` option can be either a single file or a folder. If the
+``spectrum_path`` is a folder, MS²Rescore will search for spectrum files in the directory according
+to the run names in the PSM file.
diff --git a/docs/source/userguide/output-files.rst b/docs/source/userguide/output-files.rst
index e13c238a..ee1a78f8 100644
--- a/docs/source/userguide/output-files.rst
+++ b/docs/source/userguide/output-files.rst
@@ -2,4 +2,61 @@
Output files
############
-[todo]
+Depending on the options you choose, the following files will be created. All PSMs, peptides, and
+proteins are not yet filtered at any false discovery rate (FDR) level.
+
+Main output files:
+
++-----------------------------------+----------------------------------------------------------------------------------+
+| File | Description |
++===================================+==================================================================================+
+| ``.psms.tsv`` | Main output file with rescored PSMs and their new scores |
++-----------------------------------+----------------------------------------------------------------------------------+
+| ``.report.html`` | HTML report with interactive plots showing the results and some quality control |
+| | metrics. |
++-----------------------------------+----------------------------------------------------------------------------------+
+
+Log and configuration files:
+
++--------------------------------------+--------------------------------------------------------------------------------------+
+| File | Description |
++======================================+======================================================================================+
+| ``.log.txt`` | Log file with information about the run |
++--------------------------------------+--------------------------------------------------------------------------------------+
+| ``.log.html`` | HTML version of the log file |
++--------------------------------------+--------------------------------------------------------------------------------------+
+| ``.full-config.json`` | Full configuration file with all the parameters used |
+| | as configured in the user-provided configuration file, the command line or graphical |
+| | interface, and the default values. |
++--------------------------------------+--------------------------------------------------------------------------------------+
+| ``.feature_names.tsv`` | List of the features and their descriptions |
++--------------------------------------+--------------------------------------------------------------------------------------+
+
+Rescoring engine files:
+
++-------------------------------------------------------------+-------------------------------------------------------------+
+| File | Description |
++=============================================================+=============================================================+
+| ``..psms.txt`` | PSMs and their new scores at PSM-level FDR. |
++-------------------------------------------------------------+-------------------------------------------------------------+
+| ``..peptides.txt`` | Peptides and their new scores at peptide-level FDR. |
++-------------------------------------------------------------+-------------------------------------------------------------+
+| ``..proteins.txt`` | Proteins and their new scores at protein-level FDR. |
++-------------------------------------------------------------+-------------------------------------------------------------+
+| ``..decoy.psms.txt`` | Decoy PSMs and their new scores at PSM-level FDR. |
++-------------------------------------------------------------+-------------------------------------------------------------+
+| ``..decoy.peptides.txt`` | Decoy peptides and their new scores at peptide-level FDR. |
++-------------------------------------------------------------+-------------------------------------------------------------+
+| ``..decoy.proteins.txt`` | Decoy proteins and their new scores at protein-level FDR. |
++-------------------------------------------------------------+-------------------------------------------------------------+
+| ``..weights.txt`` | Feature weights, showing feature usage in the rescoring run |
++-------------------------------------------------------------+-------------------------------------------------------------+
+
+If no rescoring engine is selected (or if Percolator was selected), the following files will also
+be written:
+
++-------------------------------------------------------------+-----------------------------------------------------------+
+| File | Description |
++=============================================================+===========================================================+
+| ``.pin`` | PSMs with all features for rescoring |
++-------------------------------------------------------------+-----------------------------------------------------------+
diff --git a/ms2rescore/__init__.py b/ms2rescore/__init__.py
index 2bac7bbe..5638cf9c 100644
--- a/ms2rescore/__init__.py
+++ b/ms2rescore/__init__.py
@@ -1,6 +1,6 @@
"""MS²Rescore: Sensitive PSM rescoring with predicted MS² peak intensities and RTs."""
-__version__ = "3.0.0b5"
+__version__ = "3.0.0"
from warnings import filterwarnings
diff --git a/ms2rescore/feature_generators/ionmob.py b/ms2rescore/feature_generators/ionmob.py
index e55e6cc6..63ed3b87 100644
--- a/ms2rescore/feature_generators/ionmob.py
+++ b/ms2rescore/feature_generators/ionmob.py
@@ -1,6 +1,20 @@
-import contextlib
+"""
+``ionmob`` collisional cross section (CCS)-based feature generator.
+
+``ionmob`` is a predictor for peptide collisional cross sections (CCS), as measured in ion mobility
+devices, such as the Bruker timsTOF instruments. More info can be found on the
+`ionmob GitHub page `_.
+
+If you use ``ionmob`` in your work, please cite the following publication:
+
+.. epigraph::
+ Teschner, D. et al. Ionmob: a Python package for prediction of peptide collisional
+ cross-section values. *Bioinformatics* 39, btad486 (2023).
+ `doi:10.1093/bioinformatics/btad486 `_
+
+"""
+
import logging
-import os
from itertools import chain
from pathlib import Path
from typing import Dict, Optional
diff --git a/ms2rescore/rescoring_engines/percolator.py b/ms2rescore/rescoring_engines/percolator.py
index da5811d5..5f7d4e5d 100644
--- a/ms2rescore/rescoring_engines/percolator.py
+++ b/ms2rescore/rescoring_engines/percolator.py
@@ -52,12 +52,13 @@ def rescore(
Aside from updating the PSM ``score``, ``qvalue``, and ``pep`` values, the following output
files are written:
- - Target PSMs: ``{output_file_root}_target_psms.pout``
- - Decoy PSMs: ``{output_file_root}_decoy_psms.pout``
- - Target Peptides: ``{output_file_root}_target_peptides.pout``
- - Decoy Peptides: ``{output_file_root}_decoy_peptides.pout``
- - Target Proteins: ``{output_file_root}_target_proteins.pout``
- - Decoy Proteins: ``{output_file_root}_decoy_proteins.pout``
+ - Target PSMs: ``{output_file_root}.percolator.psms.pout``
+ - Target peptides: ``{output_file_root}.percolator.peptides.pout``
+ - Target proteins: ``{output_file_root}.percolator.proteins.pout``
+ - Decoy PSMs: ``{output_file_root}.percolator.decoy.psms.pout``
+ - Decoy peptides: ``{output_file_root}.percolator.decoy.peptides.pout``
+ - Decoy proteins: ``{output_file_root}.percolator.decoy.proteins.pout``
+ - Feature weights: ``{output_file_root}.percolator.weights.tsv``
Percolator is run through its command line interface. Percolator must be installed separately
and the ``percolator`` command must be available in the PATH for this module to work.
@@ -79,13 +80,13 @@ def rescore(
"""
percolator_kwargs = {
- "results-psms": output_file_root + "_target_psms.pout",
- "decoy-results-psms": output_file_root + "_decoy_psms.pout",
- "results-peptides": output_file_root + "_target_peptides.pout",
- "decoy-results-peptides": output_file_root + "_decoy_peptides.pout",
- "results-proteins": output_file_root + "_target_proteins.pout",
- "decoy-results-proteins": output_file_root + "_decoy_proteins.pout",
- "weights": output_file_root + ".weights",
+ "results-psms": output_file_root + ".percolator.psms.pout",
+ "decoy-results-psms": output_file_root + ".percolator.decoy.psms.pout",
+ "results-peptides": output_file_root + ".percolator.peptides.pout",
+ "decoy-results-peptides": output_file_root + ".percolator.decoy.peptides.pout",
+ "results-proteins": output_file_root + ".percolator.proteins.pout",
+ "decoy-results-proteins": output_file_root + ".percolator.decoy.proteins.pout",
+ "weights": output_file_root + ".percolator.weights.tsv",
"verbose": LOG_LEVEL_MAP[log_level],
"num-threads": processes,
"post-processing-tdc": True,
@@ -110,9 +111,20 @@ def rescore(
logger.debug(f"Running percolator command {' '.join(percolator_cmd)}")
try:
output = subprocess.run(percolator_cmd, capture_output=True)
- except subprocess.CalledProcessError:
+ except FileNotFoundError as e:
+ if subprocess.getstatusoutput("percolator")[0] != 0:
+ raise MS2RescoreError(
+ "Could not run Percolator. Please ensure that the program is installed and "
+ "available in your PATH. See "
+ "https://ms2rescore.readthedocs.io/en/latest/installation/#installing-percolator "
+ "for more information."
+ ) from e
+ else:
+ logger.warn(f"Running Percolator resulted in an error:\n{output.stdout}")
+ raise MS2RescoreError("Percolator error") from e
+ except subprocess.CalledProcessError as e:
logger.warn(f"Running Percolator resulted in an error:\n{output.stdout}")
- raise MS2RescoreError("Percolator error")
+ raise MS2RescoreError("Percolator error") from e
logger.info(
"Percolator output: \n" + _decode_string(output.stderr), extra={"highlighter": None}
@@ -191,12 +203,3 @@ def _decode_string(encoded_string):
pass
else:
raise MS2RescoreError("Could not infer encoding of Percolator logs.")
-
-
-def _validate_cli_dependency(command):
- """Validate that command returns zero exit status."""
- if subprocess.getstatusoutput(command)[0] != 0:
- raise MS2RescoreError(
- f"Could not run command '{command}'. Please ensure that the program is installed and "
- "available in your PATH."
- )