nim Composition Space Optimization is a high-performance tool implementing several methods for selecting components (data dimensions) in compositional datasets, which optimize the data availability and density for applications such as machine learning (ML) given a constraint on the number of components to be selected, so that they can be designed in a way balancing their accuracy and domain of applicability. Making said choice is a combinatorically hard problem when data is composed of a large number of independent components due to the interdependency of components being present. Thus, efficiency of the search becomes critical for any application where interaction between components is of interest in a modeling effort, ranging:
- from market economics,
- through medicine where drug interactions can have a significant impact on the treatment,
- to materials science, where the composition and processing history are critical to resulting properties.
We are particularily interested in the latter case of materials science, where we utilize nimCSO to optimize ML deployment over our datasets on Compositionally Complex Materials (CCMs)
which are largest ever collected (from almost 550 publications) spanning up to 60 dimensions and developed within the ULTERA Project (ultera.org) carried under the
US DOE ARPA-E ULTIMATE program which aims to develop
a new generation of ultra-high temperature materials for aerospace applications, through generative machine learning models 10.20517/jmi.2021.05
driving thermodynamic modeling and experimentation 10.2139/ssrn.4689687.
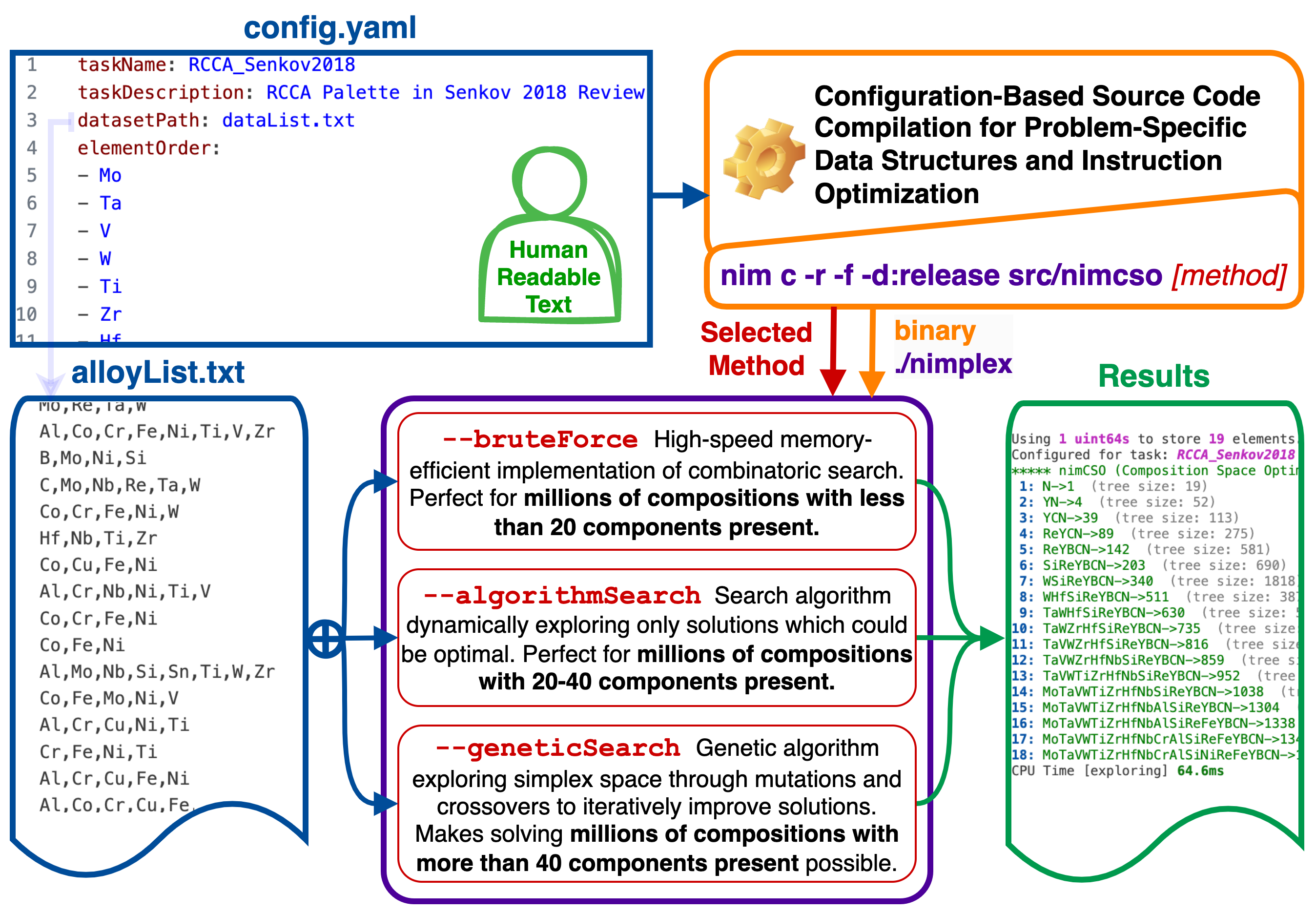
At its core, nimCSO leverages the metaprogramming ability of the Nim language to optimize itself at the compile time, both in terms of speed and memory handling,
to the specific problem statement and dataset at hand based on a human-readable configuration file. As demonstrated later in benchamrks, nimCSO reaches the physical limits of the hardware
(L1 cache latency) and can outperform an efficient native Python implementation over 400 times in terms of speed and 50 times in terms of memory usage (not counting interpreter), while
also outperforming NumPy implementation 35 and 17 times, respectively, when checking a candidate solution.
nimCSO is designed to be both (1) a user-ready tool (see figure above), implementing:
- Efficient brute force approaches (for handling up to 25 dimensions)
- Custom search algorithm (for up to 40 dimensions)
- Genetic algorithm (for any dimensionality)
and (2) a scaffold for building even more elaborate methods in the future, including heuristics going beyond
data availability. All configuration is done with a simple human-readable YAML config file and plain text data files, making it easy to modify the search method and its parameters with
no knowledge of programming and only basic command line skills. A single command is used to recompile (nim c -f) and run (-r) problem (-d:configPath=config.yaml) with nimCSO
(src/nimcso) using one of several methods. Advanced users can also quickly customize the provided methods with brief scripts using the nimCSO as a data-centric library.
To use nimCSO you don't even need to install anything, as long as you have a (free) GitHub account, since we prepared a pre-configured Codespace for you! Simply click on the link below and it will create a cloud development environment for you, with all dependencies installed for you through Conda and Nimble package managers. You can then run nimCSO through terminal or play with a Jupyter notebook we prepared.
Note: If you want to install nimCSO yourself, follow the instructions in the Installation section.
The config.yaml file is the critical component which defines several required parameters listed below. You can either just change the values in the provided config.yaml or create a custom one, like the config_rhea.yaml, and point to it at the compilation with -d:configPath=config_rhea.yaml flag. Inside, you will need to define the following parameters:
-
taskName - A
stringwith the name of the task. It does not affect the results in any way, except for being printed during runtime for easier identification. -
taskDescription - A
stringwith the description of the task. It does not affect the results in any way, except for being printed during runtime for easier identification. -
datasetPath - A
stringwith the path (relative to CWD) with the dataset file. Please see Dataset files below for details on its content. -
elementOrder - A list of
strings with the names of the elements in the dataset. The order does not affect the results in any way, except for the order in which the elements will be printed in the resulting solutions. It does determine the order in which they are stored internally though, so if you are an advanced user and, e.g., write a custom heuristic, you may want to take advantage of this to, e.g., assign a list of weights to the elements.
We wanted to make creating the input dataset as simple and as cross-platform as possible, thus the dataset file should be plain text file containing one set of elements (in any order) per line separated by commas. You can use .txt or .csv file extensions interchangeably, with no effect on the nimCSO behavior, but note that editing CSV with Excel in some countries (e.g., Italy) may cause issues. The dataset should not contain any header. The dataset can contain any elements, as the one not present in the elementOrder will be ignored at the parsing stage. It will generally look like:
Al,Cr,Hf,Mo,Ni,Re,Ru,Si,Ta,Ti,W
Al,Co,Cr,Cu,Fe,Ni,Ti
Al,B,C,Co,Cr,Hf,Mo,Ni,Ta,Ti,W,Zr
Mo,Nb,Si,Ta,W
Co,Fe,Mo,Ni,V
Hf,Nb,Ta,Ti,Zr
Mo,Nb,Ta,V,W
Al,Co,Cr,Fe,Ni,Si,Ti
Al,Co,Cr,Cu,Fe,Ni
you are also welcome to align the elements in columns, like below,
Al, B, Co, Cr
B, Cr, Fe, Ni
Al, Co, Fe, Ni
but having empty fields is not allowed, so Al, ,Co,Cr, , ,W would not be parsed correctly.
The dataset provided by default with nimCSO comes from a snapshot of the ULTERA Database and lists elements in "aggregated" alloys, which means every entry corresponds to a unique HEA composition-processing-structure triplet (which has from one to several attached properties). The dataset access is currently limited, but once published, you will be able to obtain it (and newer versions) with Python code like this using the pymongo library:
collection = client['ULTERA']['AGGREGATED_Jul2023']
elementList = [e['material']['elements'] for e in collection.find({
'material.nComponents': {'$gte': 3},
'metaSet.source': {'$in': ['LIT', 'VAL']},
'properties.source': {'$in': ['EXP', 'DFT']}
})]If you want to use nimCSO on your machine (local or remote), the best course of action is likely to install dependencies and clone the software so that you can get a ready-to-use setup you can also customize. You can do it fairly easily in just a couple of minutes.
First, you need to install Nim language compiler which on most Unix (Linux/MacOS) systems is very straightforward.
-
On MacOS, assuming you have Homebrew installed, simply:
brew install nim
-
Using
conda,miniconda,mamba, ormicromambacross-platform package manager:conda install -c conda-forge nim
-
On most Linux distributions, you should also be able to use your built-in package manager like
pacman,apt,yum, orrpm; however, the default channel/repository, especially on enterprise systems, may have an unsupported version (nim<2.0). While we do testnimCSOwith1.6versions too, your experience may be degraded, so you may want to update it or go with another option. -
You can, of course, also build it yourself from
nimsource code! It is relatively straightforward and fast compared to many other languages.
On Windows, you may consider using WSL, i.e., Windows Subsystem for Linux, which is strongly recommended, interplays well with VS Code, and will let you act as if you were on Linux. If you need to use Windows directly, you can follow these installation instructions.
Then, you can use the bundled Nimble tool (package manager for Nim, similar to Rust's crate or Python's pip) to install two top-level nim dependencies:
- arraymancer, which is a powerful N-dimensional array library, and
- yaml which parses the configuration files.
It's a single command:
nimble install --depsOnlyor, explicitly:
nimble install -y arraymancer yamlFinally, you can clone the repository and compile the library with:
git clone https://github.com/amkrajewski/nimcso
cd nimcso
nim c -r -f -d:release src/nimcsowhich will compile the library and print out concise help message with available CLI options.
And now, you are ready to use nimCSO :)
-
In general,
nimblecould give you a very similar experience topipand allow you to installnimCSOfromnimbleindex (PyPI equivalent), without manual cloning and compilation. The reason above README undergoes such additional gymnastics and places the binary in the localsrcrather than some more general location, is thatnimCSOis meant to be compiled with local YAML config files on a per-task basis, taking advantage of optimizations enabled by knowing task specifics. Thus, having it installed would, I think, confuse users and perhaps leave unnecessary files behind after task completion. -
If you must use
nim<2.0for any reason, you may want to manually install package versions known to work withnim=1.6.xusingnimble install -y [email protected] [email protected]. -
You can use
nimble list -ito verify that Nim packages were installed correctly. -
Please note that
nimused bynimCSOis the nim programming language, not apythonpackage. Whilecondawill work perfectly on Unix systems, you cannot install it withpip install nim, as thenimPython package on PyPI is an entirely different thing (an obscure interface to the Network Interface Monitor).
-
We explicitly welcome unsolicited user feedback and feature requests submitted through GitHub Issues.
-
If you wish to contribute to the development of
nimCSOyou are very welcome to do so by forking the repository and creating a pull request. As of Summer 2024, we are actively developing the code and using it in two separate research projects, so we should get back to you within a week or two. -
We are particularly interested in:
- Performance improvements, even if marginal.
- Additional I/O file format handling, like HDF5, especially on the input data side.
- Additional genetic algorithms, ideally, outperforming the current ones.
- Additional test databases and configurations. We would love to see some high entropy ceramics, glasses, and metallic glasses on the materials science side, complex microbial communities on the biology side, and polypharmacy-related data on the pharmaceutical side.
-
We are also open to helping you run our code in non-profit or academic research cases! Please do not hesitate to contact us through the GitHub issues or by email.
-
We do not enforce any strict style convention for
nimCSOcontributions, as long as code maintains high readability and overall quality. -
If you are unsure on what style to use, consult
nimcompiler style convention and try to stick to it. In general, style conventions innimlanguage are a very tricky subject compared to most languages. It is explicitly designed to not have code style conventions, even on the basic level like naming, and makes programmers read code closer to how it will be parsed into AST. The result is that collaborative projects use camelCase in one file to define a function and then kebab-case in another one to call it. Surprisingly to some,nimprogrammers tend to cherish that and even the largest projects likeArraymancerallow it.