This repo is for our NeurIPS 2023 paper DiffBP for molecule generation.
Code: Code is avaiable in [CBGBench].
Authors: Haitao Lin, Yufei Huang, Meng Liu, Xuanjing Li, Shuiwang Ji, Stan Z. Li.
Generating molecules that bind to specific proteins is an important but challenging task in drug discovery. Previous works usually generate atoms in an auto-regressive way, where element types and 3D coordinates of atoms are generated one by one. However, in real-world molecular systems, the interactions among atoms in an entire molecule are global, leading to the energy function pair-coupled among atoms. With such energy-based consideration, the modeling of probability should be based on joint distributions, rather than sequentially conditional ones. Thus, the unnatural sequentially auto-regressive modeling of molecule generation is likely to violate the physical rules, thus resulting in poor properties of the generated molecules. In this work, a generative diffusion model for molecular 3D structures based on target proteins as contextual constraints is established, at a full-atom level in a non-autoregressive way. Given a designated 3D protein binding site, our model learns the generative process that denoises both element types and 3D coordinates of an entire molecule, with an equivariant network. Experimentally, the proposed method shows competitive performance compared with prevailing works in terms of high affinity with proteins and appropriate molecule sizes as well as other drug properties such as drug-likeness of the generated molecules.
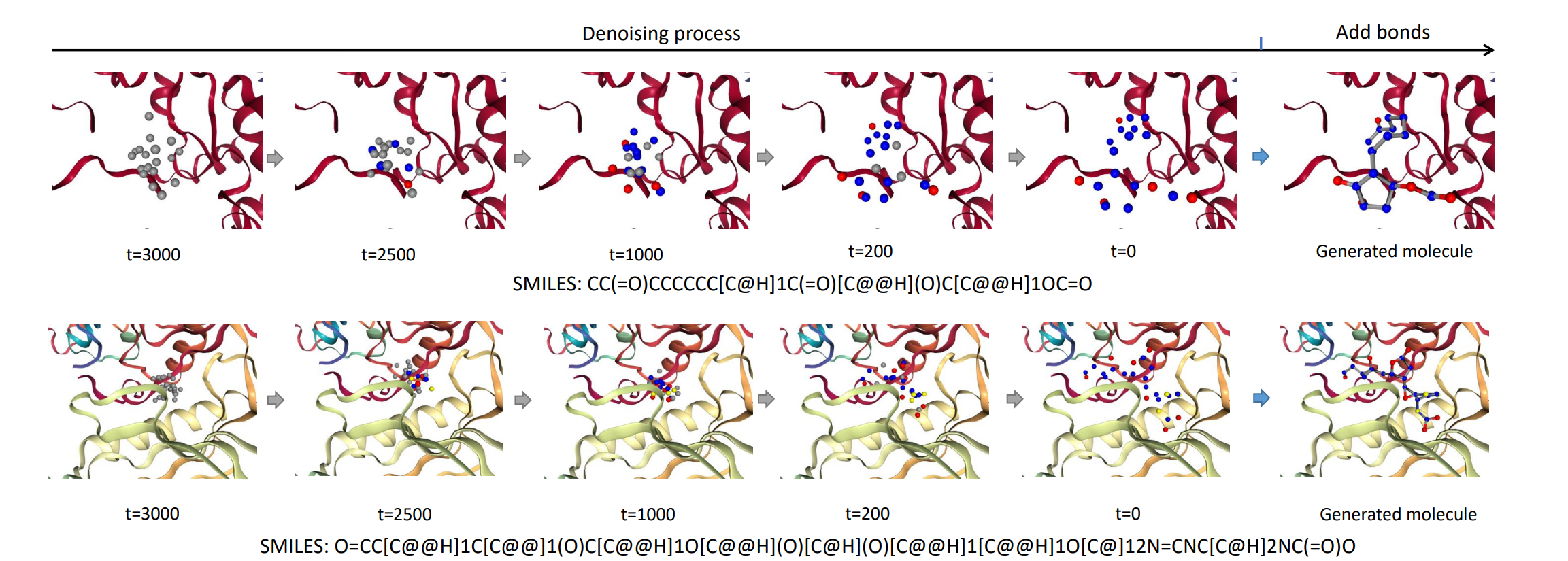
DiffBP codes have been released to [CBGBench].
If you find this work useful in your research, please consider citing:
@misc{lin2022diffbp,
title={DiffBP: Generative Diffusion of 3D Molecules for Target Protein Binding},
author={Haitao Lin and Yufei Huang and Meng Liu and Xuanjing Li and Shuiwang Ji and Stan Z. Li},
year={2022},
eprint={2211.11214},
archivePrefix={arXiv},
primaryClass={id='q-bio.BM' full_name='Biomolecules' is_active=True alt_name=None in_archive='q-bio' is_general=False description='DNA, RNA, proteins, lipids, etc.; molecular structures and folding kinetics; molecular interactions; single-molecule manipulation.'}
}