-
Notifications
You must be signed in to change notification settings - Fork 196
Multilabel DeepEdit
Multilabel DeepEdit generalizes the DeepEdit App. This means it addresses the single and multilabel segmentation tasks. Similar to DeepEdit, this App combines the power of two models in one single architecture: automatic inference, as a standard segmentation method (i.e. UNet), and interactive segmentation using clicks as the DeepGrow.
Similar to the single label DeepEdit, the training process of the multilabel DeepEdit App involves a combination of simulated clicks and standard training. As shown in the next figure, the input of the network is a concatenation of N tensors: image, and one channel per label including background; those channels contain the simulated points or clicks. This model has two types of training: For some iterations, tensors representing the clicks for each label are zeros and for the other part of the iterations, clicks are simulated so the model can receive inputs for interactive segmentation. For the clicks simulation, we developed new DeepGrow transforms that support multilabel click simulation.

To train this model, we used the Beyond the Cranial Vault Challenge dataset, which is composed of 30 images each one with 13 labels:
- spleen
- right kidney
- left kidney
- gallbladder
- esophagus
- liver
- stomach
- aorta
- inferior vena cava
- portal vein and splenic vein
- pancreas
- right adrenal gland
- left adrenal gland
If you would like to train the multilabel deepedit App on the BTCV dataset or any other dataset, you should first define the dictionary label_names in the main file before you start the training process.
For demo purposes, we trained the multilabel DeepEdit App using the DynUnet and the UNETR network on 7 organs:
label_names = { "spleen": 1, "right kidney": 2, "left kidney": 3, "liver": 6, "stomach": 7, "aorta": 8, "inferior vena cava": 9, "background": 0, }
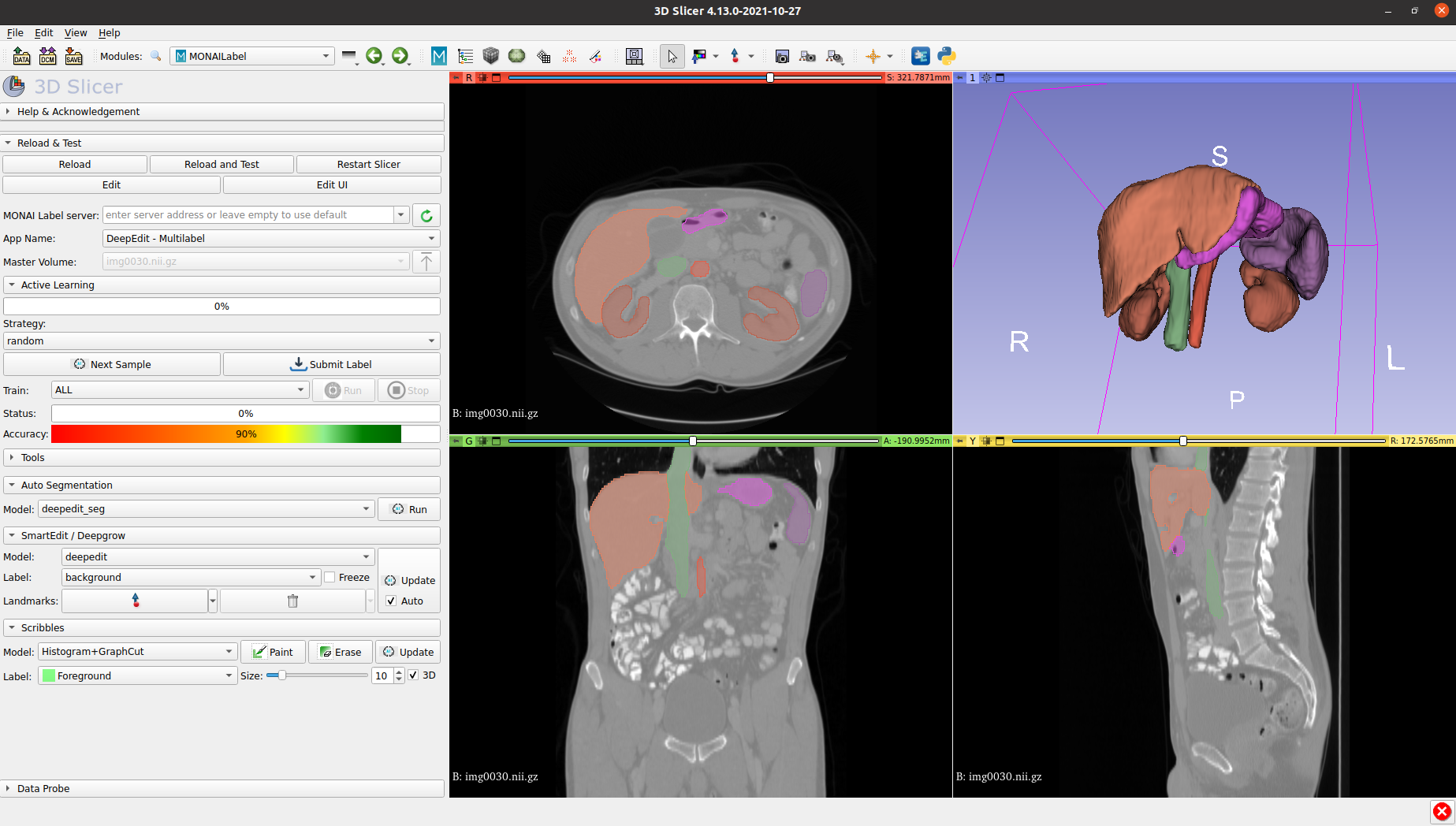
The next image shows a sample result obtained using the multilabel DeepEdit App and DynUNet as backbone:
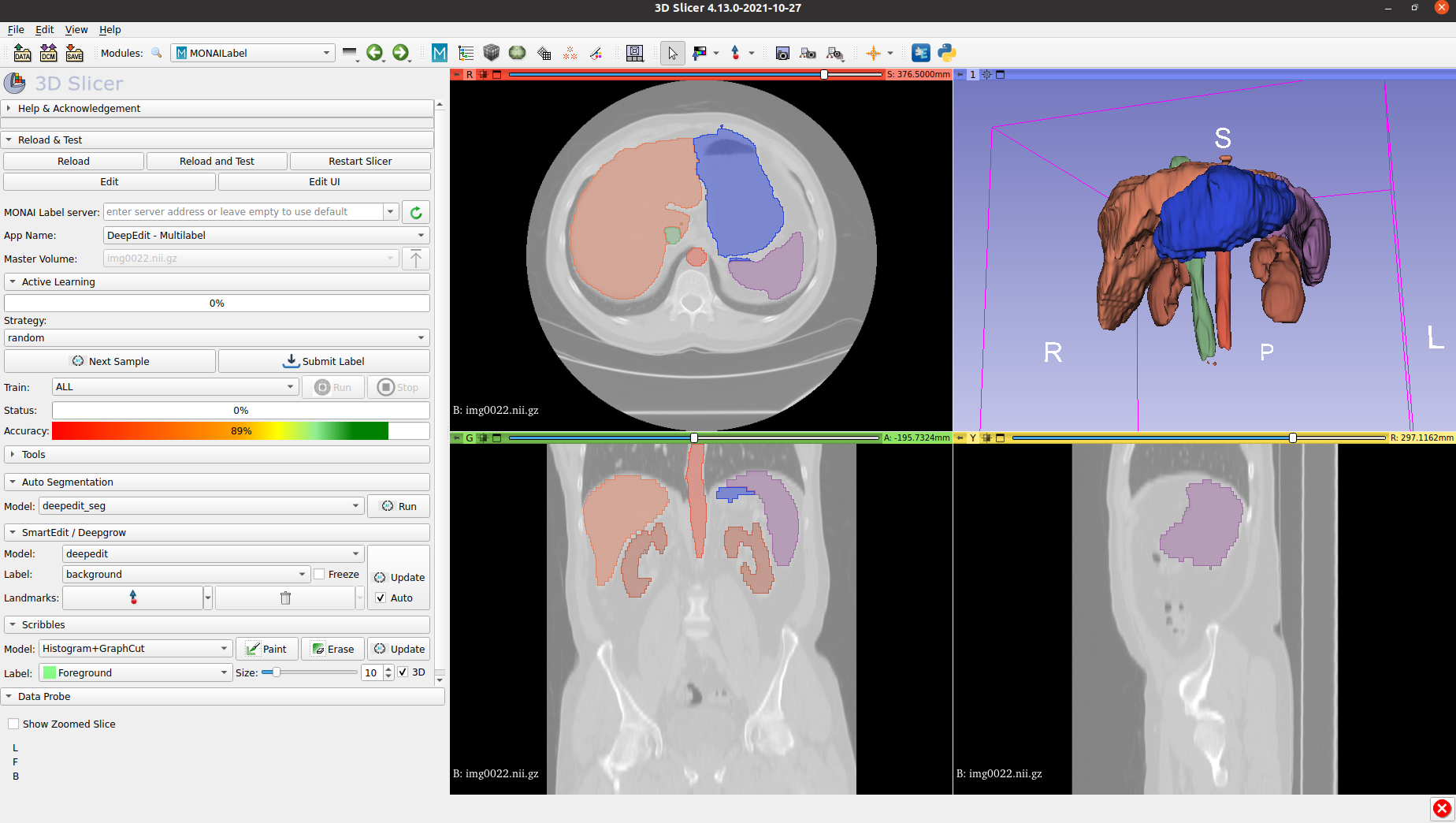
The next image shows a sample result obtained using the multilabel DeepEdit App and UNETR as backbone:
If the user wants to start this App, please choose whether to use DynUNet or UNETR as backbone:
- For DynUnet network:
monailabel start_server -a /PATH_TO_APPS/radiology/ -s /PATH_TO_DATASET/ --conf models deepedit
- For UNETR network:
monailabel start_server -a /PATH_TO_APPS/radiology/ -s /PATH_TO_DATASET/ --conf models deepedit --conf network unetr