-
Notifications
You must be signed in to change notification settings - Fork 12
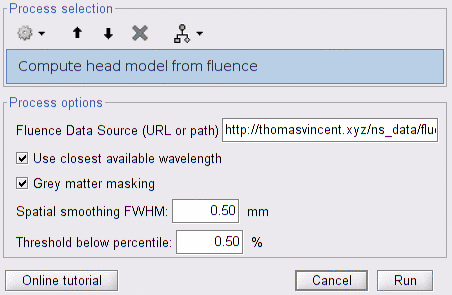
Compute head model from fluence
A head model describes how a given montage "illuminates" the brain. The term "head model" comes from the MEG/EEG literature, however the term "lead field matrix" is more often used in the NIRS setting.
This matrix is of size nb_pairs x nb_wavelength x nb_vertices and comprises the sensitivity of each cortical vertex with respect to a given optode pair at a given wavelength. For a given pair of source-detector, the sensitivity is proportional to the amount of light emitted by the source that is able to reach a given cortical point and scattered back to the detector. This sensitivity value is the product of the source fluence and the detector fluence (up to a normalization factor).
It is involved in the forward model that links cortical hemodynamic signals to the optical signals measured by the montage: y_scalp(lambda) = M(lambda) * y_cortex(lambda).
The process "Compute head model from fluence" computes the lead field matrix based on a given montage and pre-computed fluences.
-
NIRS data (montage definition): on which the process is applied (to dragg-and-dropp in the process panel).
-
5-layers segmentation:
The segmentation volume used to compute fluence data must be available in the subject anatomy folder and labeled "segmentation_5tissue". It is a volume of tissue indexes (integer from 0 to 4) with the same shape as the MRI, typically produced by freesurfer.
-
fluences computed at each vertex in the field of view:
Fluence volumes can be computed for a given anatomy using refer to .... Alternatively, if you are using a nirstorm anatomical template (eg Colin27_4NIRS), precomputed fluences are available on-line and can be downloaded automatically by this process.
Warning: fluence data take a lot of disk space (~1Mb per vertex, ~6Gb for a whole cortex - per wavelength). However, the download process will only fetch the fluence data that are required for the input montage. Expect between 200 Mb -- 500 Mb of fluence data for a typical NIRS montage.
-
Fluence Data Source: the location of the fluence data files, which can either be.
- the output path of the nirstorm processes "Compute fluence from montage" [TODO: link to doc]. This process must have been run using to the same current default anatomy.
- the URL of nirstorm repository: fluence data precomputed on the anatomical templates (eg Colin27_4NIRS).
-
Use closest available wavelength: if fluences for a given wavelength are not found, then use fluences of the closest available wavelength in the data source. We recommend using the exact wavelength. If an anatomical template is used and a precomputation is not available for a given wavelength, then it is acceptable to use an inexact wavelength. The difference will be negligible compared with the anatomical uncertainty.
-
Grey matter masking: constrain fluence data to the grey matter, to avoid partial volume effects.
-
Spatial smoothing FWHM: full width at half maximum of the spatial smoothing. If 0, then smoothing is not applied.
-
Threshold (% of max-min): Set to 0 the sensitivity values below the given percentage of value amplitude. The threshold is computed as
min + thresh_%/100 * (max - min), wheremaxandminare evaluated over sensitivity values for all pairs and wavelengths. If this parameter is 0, then no thresholding is applied. -
** Use all possible pairs **: use all combinations of source/detector instead of actual montage. This is useful in the case of multiple subject acquisitions, where pairings vary but the optode coordinates remain the same. It happens when filtering channels based on measured signals: some subjects may discard some pairs while others may keep them. To save time, compute the head model on all pairs once and then use process "Extract sub head model" [TODO: add link].
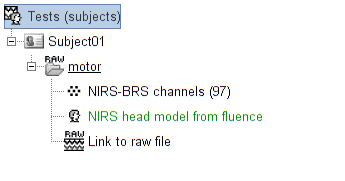
The process creates a head model brainstorm object (in green in the above figure), that can be used for source reconstruction or signal projection.
Sensitivity maps can be extracted from the head model as brainstorm cortical functional mappings. To do so, drag-and-drop the same NIRS data on which the head model was computed, then use the process "Extract sensitivity surfaces from head model".
It will produce:
- the sum of all sensitivity values across pairs, for each wavelength
- the sensitivity maps for each pair and each wavelength.
Here is an example produced on some motor data:
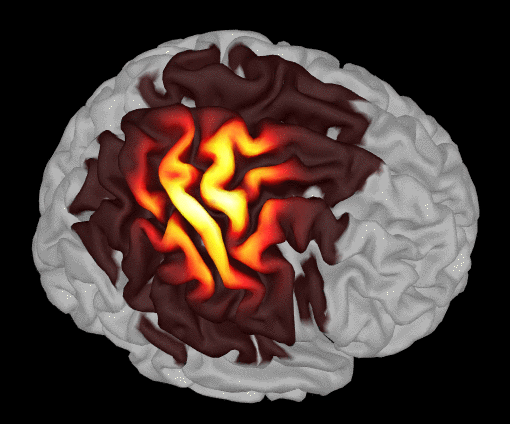
The following image shows the summed sensitivity map for wavelength 685nm:
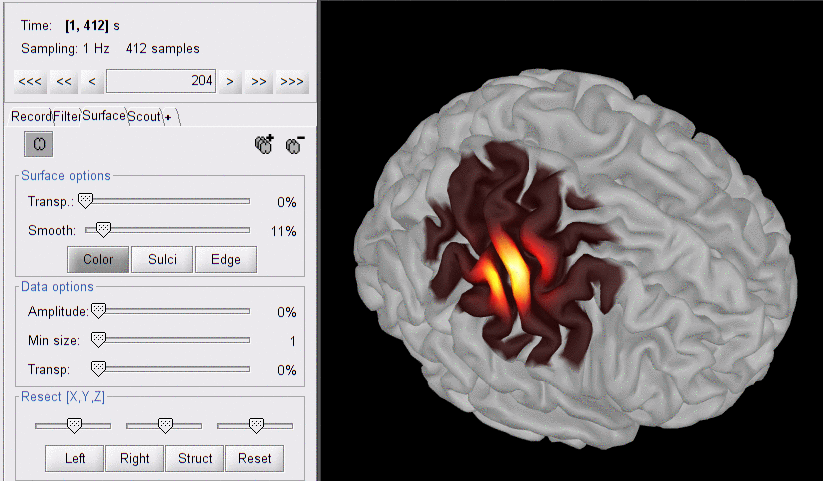
For the pairwise sensitivity maps, we used a trick to store pair information in the temporal axis. The two first digits of the time value (from the right) correspond to the detector index and the remaining digits to the source index. So time=101 corresponds to S1D1. With the same logic, time=210 is used for S2D10. Note that some time indexes do not correspond to any source-detector pair and the sensitivity map is then empty at these positions.
Here is the sensitivity map for source S2 and detector D4 (note the temporal marker at 204 s):
-
Tutorials:
-
Workshop and courses:
-
Manual:
-
Contribute: